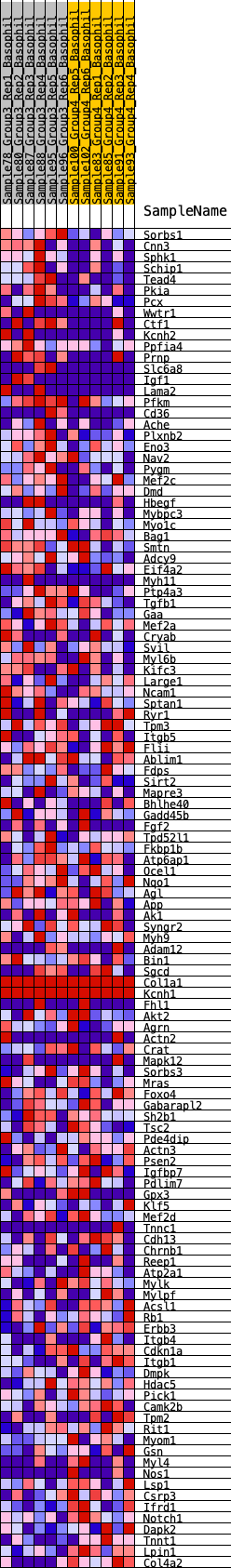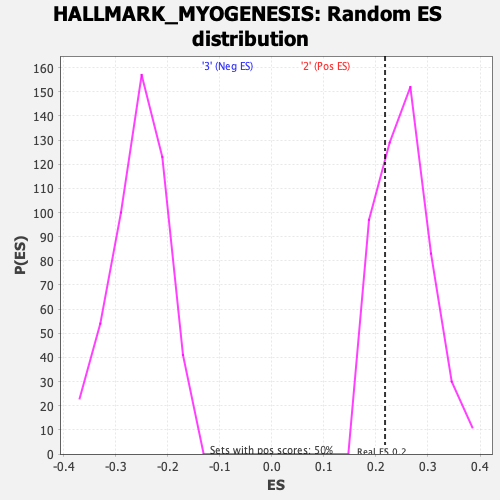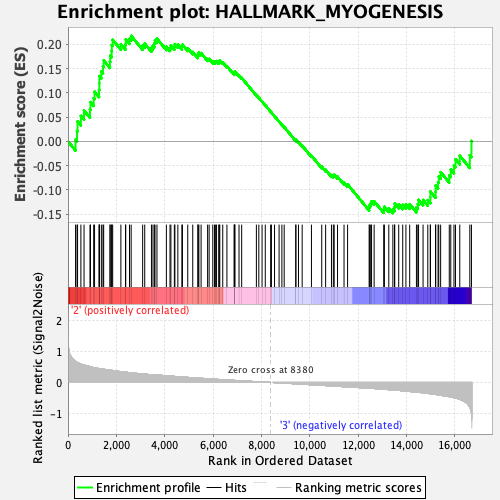
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.21744777 |
| Normalized Enrichment Score (NES) | 0.85314965 |
| Nominal p-value | 0.7211155 |
| FDR q-value | 0.9089881 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sorbs1 | 312 | 0.684 | 0.0038 | Yes |
| 2 | Cnn3 | 373 | 0.646 | 0.0215 | Yes |
| 3 | Sphk1 | 395 | 0.636 | 0.0412 | Yes |
| 4 | Schip1 | 533 | 0.594 | 0.0525 | Yes |
| 5 | Tead4 | 663 | 0.567 | 0.0635 | Yes |
| 6 | Pkia | 909 | 0.510 | 0.0655 | Yes |
| 7 | Pcx | 930 | 0.505 | 0.0810 | Yes |
| 8 | Wwtr1 | 1062 | 0.478 | 0.0889 | Yes |
| 9 | Ctf1 | 1098 | 0.474 | 0.1024 | Yes |
| 10 | Kcnh2 | 1284 | 0.447 | 0.1060 | Yes |
| 11 | Ppfia4 | 1297 | 0.445 | 0.1200 | Yes |
| 12 | Prnp | 1299 | 0.445 | 0.1346 | Yes |
| 13 | Slc6a8 | 1380 | 0.437 | 0.1442 | Yes |
| 14 | Igf1 | 1446 | 0.429 | 0.1545 | Yes |
| 15 | Lama2 | 1475 | 0.426 | 0.1668 | Yes |
| 16 | Pfkm | 1735 | 0.397 | 0.1643 | Yes |
| 17 | Cd36 | 1746 | 0.395 | 0.1768 | Yes |
| 18 | Ache | 1805 | 0.390 | 0.1862 | Yes |
| 19 | Plxnb2 | 1812 | 0.389 | 0.1987 | Yes |
| 20 | Eno3 | 1848 | 0.386 | 0.2093 | Yes |
| 21 | Nav2 | 2187 | 0.347 | 0.2004 | Yes |
| 22 | Pygm | 2377 | 0.331 | 0.1999 | Yes |
| 23 | Mef2c | 2388 | 0.331 | 0.2102 | Yes |
| 24 | Dmd | 2544 | 0.315 | 0.2113 | Yes |
| 25 | Hbegf | 2612 | 0.310 | 0.2174 | Yes |
| 26 | Mybpc3 | 3087 | 0.271 | 0.1978 | No |
| 27 | Myo1c | 3173 | 0.265 | 0.2014 | No |
| 28 | Bag1 | 3451 | 0.249 | 0.1930 | No |
| 29 | Smtn | 3514 | 0.247 | 0.1974 | No |
| 30 | Adcy9 | 3572 | 0.245 | 0.2020 | No |
| 31 | Eif4a2 | 3597 | 0.243 | 0.2086 | No |
| 32 | Myh11 | 3679 | 0.238 | 0.2116 | No |
| 33 | Ptp4a3 | 4068 | 0.214 | 0.1952 | No |
| 34 | Tgfb1 | 4218 | 0.203 | 0.1929 | No |
| 35 | Gaa | 4255 | 0.202 | 0.1974 | No |
| 36 | Mef2a | 4398 | 0.195 | 0.1953 | No |
| 37 | Cryab | 4420 | 0.194 | 0.2004 | No |
| 38 | Svil | 4543 | 0.187 | 0.1992 | No |
| 39 | Myl6b | 4709 | 0.176 | 0.1951 | No |
| 40 | Kifc3 | 4733 | 0.174 | 0.1995 | No |
| 41 | Large1 | 4952 | 0.163 | 0.1917 | No |
| 42 | Ncam1 | 5159 | 0.152 | 0.1843 | No |
| 43 | Sptan1 | 5368 | 0.141 | 0.1764 | No |
| 44 | Ryr1 | 5385 | 0.140 | 0.1801 | No |
| 45 | Tpm3 | 5412 | 0.139 | 0.1831 | No |
| 46 | Itgb5 | 5504 | 0.135 | 0.1820 | No |
| 47 | Flii | 5773 | 0.120 | 0.1699 | No |
| 48 | Ablim1 | 5836 | 0.117 | 0.1700 | No |
| 49 | Fdps | 5987 | 0.109 | 0.1645 | No |
| 50 | Sirt2 | 6057 | 0.105 | 0.1639 | No |
| 51 | Mapre3 | 6105 | 0.103 | 0.1644 | No |
| 52 | Bhlhe40 | 6143 | 0.102 | 0.1656 | No |
| 53 | Gadd45b | 6234 | 0.097 | 0.1633 | No |
| 54 | Fgf2 | 6255 | 0.096 | 0.1653 | No |
| 55 | Tpd52l1 | 6277 | 0.096 | 0.1672 | No |
| 56 | Fkbp1b | 6399 | 0.089 | 0.1628 | No |
| 57 | Atp6ap1 | 6572 | 0.079 | 0.1551 | No |
| 58 | Ocel1 | 6872 | 0.066 | 0.1392 | No |
| 59 | Nqo1 | 6879 | 0.066 | 0.1410 | No |
| 60 | Agl | 6892 | 0.065 | 0.1425 | No |
| 61 | App | 6902 | 0.065 | 0.1441 | No |
| 62 | Ak1 | 7072 | 0.057 | 0.1358 | No |
| 63 | Syngr2 | 7182 | 0.052 | 0.1309 | No |
| 64 | Myh9 | 7790 | 0.025 | 0.0951 | No |
| 65 | Adam12 | 7891 | 0.021 | 0.0898 | No |
| 66 | Bin1 | 8025 | 0.015 | 0.0823 | No |
| 67 | Sgcd | 8158 | 0.009 | 0.0746 | No |
| 68 | Col1a1 | 8380 | 0.000 | 0.0613 | No |
| 69 | Kcnh1 | 8412 | 0.000 | 0.0594 | No |
| 70 | Fhl1 | 8541 | -0.003 | 0.0518 | No |
| 71 | Akt2 | 8729 | -0.012 | 0.0409 | No |
| 72 | Agrn | 8845 | -0.017 | 0.0346 | No |
| 73 | Actn2 | 8940 | -0.021 | 0.0296 | No |
| 74 | Crat | 9420 | -0.043 | 0.0021 | No |
| 75 | Mapk12 | 9423 | -0.043 | 0.0034 | No |
| 76 | Sorbs3 | 9531 | -0.047 | -0.0015 | No |
| 77 | Mras | 9684 | -0.054 | -0.0089 | No |
| 78 | Foxo4 | 10067 | -0.072 | -0.0295 | No |
| 79 | Gabarapl2 | 10493 | -0.091 | -0.0521 | No |
| 80 | Sh2b1 | 10654 | -0.099 | -0.0585 | No |
| 81 | Tsc2 | 10895 | -0.110 | -0.0694 | No |
| 82 | Pde4dip | 10972 | -0.114 | -0.0702 | No |
| 83 | Actn3 | 11007 | -0.115 | -0.0684 | No |
| 84 | Psen2 | 11146 | -0.122 | -0.0727 | No |
| 85 | Igfbp7 | 11415 | -0.135 | -0.0845 | No |
| 86 | Pdlim7 | 11562 | -0.141 | -0.0886 | No |
| 87 | Gpx3 | 12453 | -0.185 | -0.1362 | No |
| 88 | Klf5 | 12475 | -0.186 | -0.1313 | No |
| 89 | Mef2d | 12519 | -0.189 | -0.1277 | No |
| 90 | Tnnc1 | 12553 | -0.189 | -0.1234 | No |
| 91 | Cdh13 | 12660 | -0.194 | -0.1234 | No |
| 92 | Chrnb1 | 13056 | -0.218 | -0.1400 | No |
| 93 | Reep1 | 13084 | -0.219 | -0.1344 | No |
| 94 | Atp2a1 | 13266 | -0.231 | -0.1377 | No |
| 95 | Mylk | 13434 | -0.241 | -0.1399 | No |
| 96 | Mylpf | 13503 | -0.245 | -0.1359 | No |
| 97 | Acsl1 | 13511 | -0.246 | -0.1282 | No |
| 98 | Rb1 | 13676 | -0.256 | -0.1296 | No |
| 99 | Erbb3 | 13842 | -0.266 | -0.1308 | No |
| 100 | Itgb4 | 13974 | -0.276 | -0.1296 | No |
| 101 | Cdkn1a | 14130 | -0.289 | -0.1294 | No |
| 102 | Itgb1 | 14409 | -0.309 | -0.1360 | No |
| 103 | Dmpk | 14467 | -0.314 | -0.1290 | No |
| 104 | Hdac5 | 14497 | -0.316 | -0.1204 | No |
| 105 | Pick1 | 14683 | -0.330 | -0.1206 | No |
| 106 | Camk2b | 14879 | -0.351 | -0.1208 | No |
| 107 | Tpm2 | 14984 | -0.362 | -0.1151 | No |
| 108 | Rit1 | 14986 | -0.362 | -0.1032 | No |
| 109 | Myom1 | 15201 | -0.382 | -0.1035 | No |
| 110 | Gsn | 15205 | -0.383 | -0.0911 | No |
| 111 | Myl4 | 15304 | -0.398 | -0.0838 | No |
| 112 | Nos1 | 15332 | -0.400 | -0.0723 | No |
| 113 | Lsp1 | 15410 | -0.410 | -0.0634 | No |
| 114 | Csrp3 | 15767 | -0.455 | -0.0698 | No |
| 115 | Ifrd1 | 15829 | -0.467 | -0.0581 | No |
| 116 | Notch1 | 15961 | -0.491 | -0.0498 | No |
| 117 | Dapk2 | 16023 | -0.504 | -0.0368 | No |
| 118 | Tnnt1 | 16201 | -0.541 | -0.0296 | No |
| 119 | Lpin1 | 16617 | -0.794 | -0.0285 | No |
| 120 | Col4a2 | 16684 | -1.010 | 0.0009 | No |