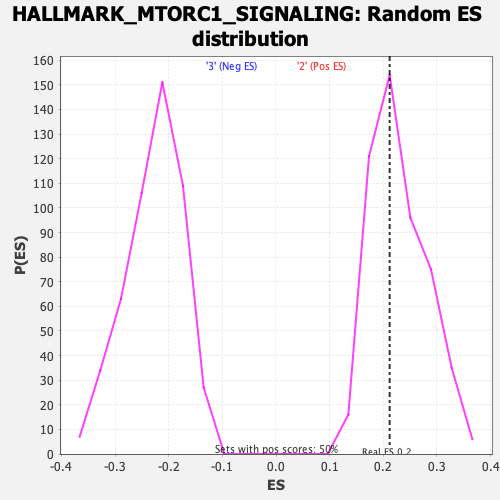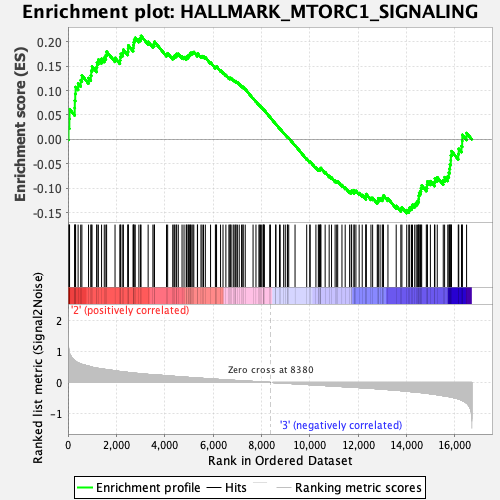
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MTORC1_SIGNALING |
| Enrichment Score (ES) | 0.21193464 |
| Normalized Enrichment Score (NES) | 0.92827135 |
| Nominal p-value | 0.5467197 |
| FDR q-value | 0.9450627 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hmbs | 22 | 1.160 | 0.0238 | Yes |
| 2 | Hmgcs1 | 56 | 0.939 | 0.0422 | Yes |
| 3 | Psat1 | 64 | 0.924 | 0.0618 | Yes |
| 4 | Insig1 | 277 | 0.706 | 0.0643 | Yes |
| 5 | Idi1 | 282 | 0.704 | 0.0793 | Yes |
| 6 | Fgl2 | 299 | 0.690 | 0.0933 | Yes |
| 7 | Sqle | 314 | 0.683 | 0.1073 | Yes |
| 8 | Ddit4 | 418 | 0.628 | 0.1147 | Yes |
| 9 | Psmd13 | 526 | 0.597 | 0.1211 | Yes |
| 10 | Nampt | 574 | 0.581 | 0.1309 | Yes |
| 11 | Bub1 | 849 | 0.524 | 0.1257 | Yes |
| 12 | Gla | 944 | 0.501 | 0.1309 | Yes |
| 13 | Cyp51 | 961 | 0.498 | 0.1407 | Yes |
| 14 | Plod2 | 994 | 0.492 | 0.1494 | Yes |
| 15 | Atp6v1d | 1185 | 0.460 | 0.1479 | Yes |
| 16 | Gtf2h1 | 1198 | 0.457 | 0.1571 | Yes |
| 17 | Aurka | 1256 | 0.452 | 0.1634 | Yes |
| 18 | Nup205 | 1388 | 0.437 | 0.1650 | Yes |
| 19 | Hspe1 | 1500 | 0.423 | 0.1674 | Yes |
| 20 | Hmgcr | 1567 | 0.417 | 0.1725 | Yes |
| 21 | Sord | 1597 | 0.413 | 0.1797 | Yes |
| 22 | Ak4 | 1945 | 0.376 | 0.1668 | Yes |
| 23 | Pgm1 | 2147 | 0.352 | 0.1623 | Yes |
| 24 | Dhcr24 | 2162 | 0.350 | 0.1690 | Yes |
| 25 | Mthfd2l | 2186 | 0.347 | 0.1752 | Yes |
| 26 | Etf1 | 2268 | 0.340 | 0.1776 | Yes |
| 27 | Hspd1 | 2289 | 0.339 | 0.1838 | Yes |
| 28 | Bcat1 | 2474 | 0.322 | 0.1796 | Yes |
| 29 | Nufip1 | 2486 | 0.322 | 0.1859 | Yes |
| 30 | Gbe1 | 2494 | 0.320 | 0.1924 | Yes |
| 31 | Psme3 | 2693 | 0.304 | 0.1870 | Yes |
| 32 | Gga2 | 2703 | 0.303 | 0.1931 | Yes |
| 33 | Dhfr | 2719 | 0.302 | 0.1987 | Yes |
| 34 | Cth | 2737 | 0.301 | 0.2042 | Yes |
| 35 | Ccnf | 2780 | 0.297 | 0.2081 | Yes |
| 36 | Tes | 2931 | 0.284 | 0.2052 | Yes |
| 37 | Ung | 2996 | 0.278 | 0.2073 | Yes |
| 38 | Psmd12 | 3020 | 0.276 | 0.2119 | Yes |
| 39 | Add3 | 3314 | 0.258 | 0.1998 | No |
| 40 | Hspa9 | 3511 | 0.247 | 0.1933 | No |
| 41 | Slc37a4 | 3560 | 0.245 | 0.1957 | No |
| 42 | Slc7a5 | 3577 | 0.245 | 0.2000 | No |
| 43 | Rdh11 | 4074 | 0.213 | 0.1746 | No |
| 44 | Pfkl | 4121 | 0.210 | 0.1764 | No |
| 45 | Immt | 4331 | 0.199 | 0.1680 | No |
| 46 | Tomm40 | 4380 | 0.196 | 0.1694 | No |
| 47 | Got1 | 4412 | 0.194 | 0.1717 | No |
| 48 | Ykt6 | 4470 | 0.191 | 0.1724 | No |
| 49 | Ppa1 | 4494 | 0.190 | 0.1751 | No |
| 50 | Polr3g | 4561 | 0.185 | 0.1751 | No |
| 51 | Cct6a | 4716 | 0.175 | 0.1696 | No |
| 52 | Tpi1 | 4798 | 0.171 | 0.1684 | No |
| 53 | Ddx39a | 4888 | 0.166 | 0.1666 | No |
| 54 | Plk1 | 4904 | 0.166 | 0.1693 | No |
| 55 | Acsl3 | 4964 | 0.163 | 0.1693 | No |
| 56 | Txnrd1 | 4977 | 0.162 | 0.1721 | No |
| 57 | Hspa4 | 5016 | 0.160 | 0.1732 | No |
| 58 | Atp2a2 | 5050 | 0.159 | 0.1747 | No |
| 59 | Psmc6 | 5068 | 0.157 | 0.1771 | No |
| 60 | Psmb5 | 5120 | 0.154 | 0.1773 | No |
| 61 | Elovl5 | 5165 | 0.151 | 0.1779 | No |
| 62 | Nupr1 | 5203 | 0.149 | 0.1789 | No |
| 63 | Acly | 5351 | 0.142 | 0.1731 | No |
| 64 | Gmps | 5357 | 0.142 | 0.1759 | No |
| 65 | Slc7a11 | 5499 | 0.135 | 0.1703 | No |
| 66 | Glrx | 5545 | 0.133 | 0.1704 | No |
| 67 | Qdpr | 5604 | 0.129 | 0.1697 | No |
| 68 | Prdx1 | 5679 | 0.125 | 0.1679 | No |
| 69 | Ifi30 | 5895 | 0.114 | 0.1574 | No |
| 70 | Wars1 | 6095 | 0.104 | 0.1476 | No |
| 71 | Ldlr | 6119 | 0.103 | 0.1484 | No |
| 72 | Bhlhe40 | 6143 | 0.102 | 0.1492 | No |
| 73 | Egln3 | 6308 | 0.094 | 0.1413 | No |
| 74 | Fads1 | 6410 | 0.088 | 0.1371 | No |
| 75 | Psmd14 | 6529 | 0.082 | 0.1318 | No |
| 76 | Srd5a1 | 6654 | 0.075 | 0.1259 | No |
| 77 | Sec11a | 6697 | 0.074 | 0.1249 | No |
| 78 | Rrm2 | 6708 | 0.073 | 0.1259 | No |
| 79 | Xbp1 | 6745 | 0.072 | 0.1253 | No |
| 80 | Slc2a1 | 6830 | 0.068 | 0.1217 | No |
| 81 | Stard4 | 6869 | 0.067 | 0.1208 | No |
| 82 | Slc2a3 | 6939 | 0.063 | 0.1180 | No |
| 83 | Fads2 | 6944 | 0.063 | 0.1191 | No |
| 84 | Gsk3b | 7004 | 0.060 | 0.1169 | No |
| 85 | Nfyc | 7081 | 0.057 | 0.1135 | No |
| 86 | Sc5d | 7177 | 0.053 | 0.1089 | No |
| 87 | Tuba4a | 7237 | 0.050 | 0.1064 | No |
| 88 | Mcm2 | 7238 | 0.050 | 0.1075 | No |
| 89 | Skap2 | 7329 | 0.046 | 0.1030 | No |
| 90 | Acaca | 7653 | 0.032 | 0.0841 | No |
| 91 | Ebp | 7773 | 0.026 | 0.0775 | No |
| 92 | Aldoa | 7895 | 0.021 | 0.0706 | No |
| 93 | Uso1 | 7928 | 0.019 | 0.0691 | No |
| 94 | Sqstm1 | 7958 | 0.018 | 0.0677 | No |
| 95 | Sdf2l1 | 8010 | 0.015 | 0.0650 | No |
| 96 | Nmt1 | 8067 | 0.013 | 0.0619 | No |
| 97 | Actr2 | 8089 | 0.012 | 0.0609 | No |
| 98 | Rab1a | 8095 | 0.012 | 0.0608 | No |
| 99 | Elovl6 | 8120 | 0.011 | 0.0596 | No |
| 100 | Mthfd2 | 8342 | 0.001 | 0.0462 | No |
| 101 | Pdap1 | 8372 | 0.000 | 0.0445 | No |
| 102 | Slc6a6 | 8586 | -0.006 | 0.0317 | No |
| 103 | Btg2 | 8599 | -0.007 | 0.0312 | No |
| 104 | Mllt11 | 8746 | -0.013 | 0.0226 | No |
| 105 | M6pr | 8775 | -0.015 | 0.0212 | No |
| 106 | Pitpnb | 8918 | -0.020 | 0.0130 | No |
| 107 | Map2k3 | 8985 | -0.023 | 0.0096 | No |
| 108 | Uchl5 | 9071 | -0.026 | 0.0050 | No |
| 109 | Cops5 | 9079 | -0.026 | 0.0051 | No |
| 110 | Psma3 | 9128 | -0.029 | 0.0028 | No |
| 111 | Mcm4 | 9389 | -0.042 | -0.0120 | No |
| 112 | Ssr1 | 9871 | -0.063 | -0.0398 | No |
| 113 | Psmg1 | 10000 | -0.069 | -0.0460 | No |
| 114 | Phgdh | 10012 | -0.069 | -0.0452 | No |
| 115 | Vldlr | 10252 | -0.080 | -0.0580 | No |
| 116 | Cd9 | 10352 | -0.085 | -0.0621 | No |
| 117 | Ddit3 | 10376 | -0.086 | -0.0616 | No |
| 118 | Actr3 | 10402 | -0.087 | -0.0612 | No |
| 119 | Adipor2 | 10440 | -0.089 | -0.0616 | No |
| 120 | Tm7sf2 | 10450 | -0.090 | -0.0601 | No |
| 121 | Rpa1 | 10455 | -0.090 | -0.0584 | No |
| 122 | Stip1 | 10634 | -0.098 | -0.0671 | No |
| 123 | Dapp1 | 10800 | -0.106 | -0.0748 | No |
| 124 | Dhcr7 | 10898 | -0.110 | -0.0783 | No |
| 125 | Canx | 11046 | -0.117 | -0.0846 | No |
| 126 | Fkbp2 | 11111 | -0.120 | -0.0859 | No |
| 127 | Lta4h | 11153 | -0.122 | -0.0857 | No |
| 128 | Cfp | 11329 | -0.131 | -0.0935 | No |
| 129 | Eif2s2 | 11463 | -0.137 | -0.0986 | No |
| 130 | Ppp1r15a | 11646 | -0.145 | -0.1065 | No |
| 131 | Rrp9 | 11713 | -0.148 | -0.1072 | No |
| 132 | Ufm1 | 11730 | -0.149 | -0.1050 | No |
| 133 | Psmc4 | 11829 | -0.154 | -0.1076 | No |
| 134 | Cdc25a | 11832 | -0.154 | -0.1043 | No |
| 135 | Calr | 11906 | -0.157 | -0.1053 | No |
| 136 | Eno1b | 12040 | -0.163 | -0.1099 | No |
| 137 | Ppia | 12167 | -0.170 | -0.1138 | No |
| 138 | Tbk1 | 12320 | -0.177 | -0.1192 | No |
| 139 | Ube2d3 | 12323 | -0.177 | -0.1154 | No |
| 140 | Sytl2 | 12328 | -0.178 | -0.1118 | No |
| 141 | Lgmn | 12528 | -0.189 | -0.1198 | No |
| 142 | Cacybp | 12598 | -0.191 | -0.1198 | No |
| 143 | Asns | 12789 | -0.202 | -0.1270 | No |
| 144 | Rpn1 | 12822 | -0.204 | -0.1245 | No |
| 145 | Itgb2 | 12829 | -0.204 | -0.1204 | No |
| 146 | Abcf2 | 12906 | -0.209 | -0.1205 | No |
| 147 | Tcea1 | 12997 | -0.214 | -0.1213 | No |
| 148 | Fdxr | 13024 | -0.216 | -0.1182 | No |
| 149 | Cyb5b | 13046 | -0.217 | -0.1147 | No |
| 150 | Pik3r3 | 13231 | -0.228 | -0.1210 | No |
| 151 | Gpi1 | 13572 | -0.248 | -0.1362 | No |
| 152 | Ero1a | 13764 | -0.261 | -0.1421 | No |
| 153 | Gsr | 13808 | -0.264 | -0.1389 | No |
| 154 | Tmem97 | 14007 | -0.279 | -0.1449 | No |
| 155 | Ctsc | 14093 | -0.286 | -0.1438 | No |
| 156 | Cdkn1a | 14130 | -0.289 | -0.1397 | No |
| 157 | Idh1 | 14205 | -0.295 | -0.1378 | No |
| 158 | Sla | 14246 | -0.299 | -0.1337 | No |
| 159 | Slc1a5 | 14338 | -0.306 | -0.1326 | No |
| 160 | Hk2 | 14400 | -0.308 | -0.1296 | No |
| 161 | Cxcr4 | 14458 | -0.313 | -0.1263 | No |
| 162 | Ldha | 14504 | -0.317 | -0.1222 | No |
| 163 | Eef1e1 | 14507 | -0.317 | -0.1154 | No |
| 164 | Arpc5l | 14522 | -0.318 | -0.1094 | No |
| 165 | P4ha1 | 14579 | -0.323 | -0.1058 | No |
| 166 | Tubg1 | 14597 | -0.324 | -0.0998 | No |
| 167 | Edem1 | 14626 | -0.326 | -0.0944 | No |
| 168 | Hspa5 | 14817 | -0.345 | -0.0984 | No |
| 169 | Nfkbib | 14848 | -0.348 | -0.0927 | No |
| 170 | Pnp | 14863 | -0.349 | -0.0860 | No |
| 171 | Rit1 | 14986 | -0.362 | -0.0855 | No |
| 172 | Shmt2 | 15155 | -0.377 | -0.0875 | No |
| 173 | Psph | 15172 | -0.379 | -0.0802 | No |
| 174 | Ccng1 | 15270 | -0.393 | -0.0776 | No |
| 175 | Pno1 | 15518 | -0.424 | -0.0833 | No |
| 176 | Coro1a | 15574 | -0.432 | -0.0773 | No |
| 177 | Gclc | 15706 | -0.450 | -0.0755 | No |
| 178 | Psma4 | 15750 | -0.453 | -0.0683 | No |
| 179 | Nfil3 | 15778 | -0.458 | -0.0600 | No |
| 180 | G6pdx | 15795 | -0.461 | -0.0509 | No |
| 181 | Ifrd1 | 15829 | -0.467 | -0.0428 | No |
| 182 | Pdk1 | 15833 | -0.468 | -0.0328 | No |
| 183 | Psmc2 | 15858 | -0.472 | -0.0240 | No |
| 184 | Tfrc | 16139 | -0.524 | -0.0296 | No |
| 185 | Hsp90b1 | 16159 | -0.530 | -0.0193 | No |
| 186 | Pgk1 | 16272 | -0.565 | -0.0138 | No |
| 187 | Niban1 | 16300 | -0.577 | -0.0029 | No |
| 188 | Slc1a4 | 16305 | -0.579 | 0.0094 | No |
| 189 | Serp1 | 16483 | -0.664 | 0.0131 | No |