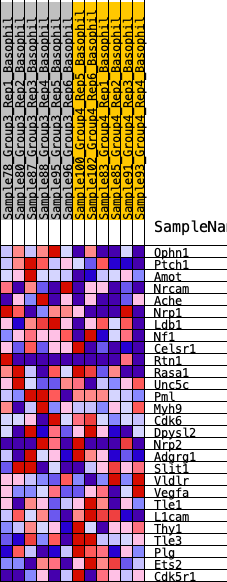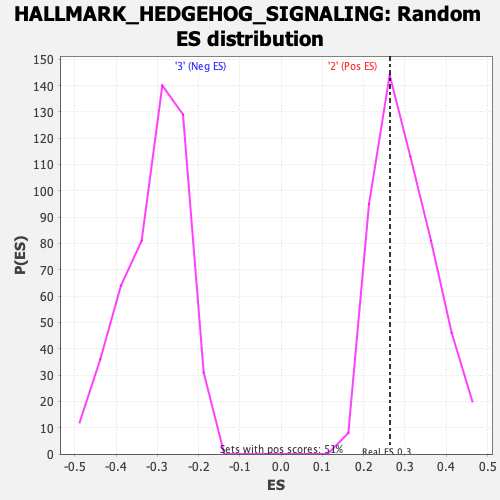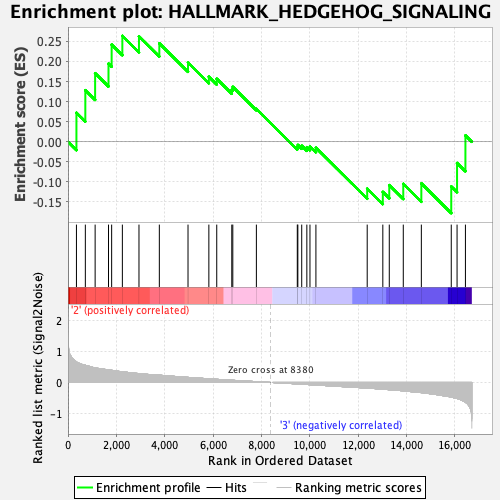
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.2638055 |
| Normalized Enrichment Score (NES) | 0.87674665 |
| Nominal p-value | 0.64102566 |
| FDR q-value | 0.90540653 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ophn1 | 349 | 0.660 | 0.0723 | Yes |
| 2 | Ptch1 | 717 | 0.553 | 0.1284 | Yes |
| 3 | Amot | 1122 | 0.471 | 0.1707 | Yes |
| 4 | Nrcam | 1675 | 0.404 | 0.1947 | Yes |
| 5 | Ache | 1805 | 0.390 | 0.2421 | Yes |
| 6 | Nrp1 | 2248 | 0.341 | 0.2638 | Yes |
| 7 | Ldb1 | 2935 | 0.284 | 0.2628 | No |
| 8 | Nf1 | 3777 | 0.233 | 0.2453 | No |
| 9 | Celsr1 | 4963 | 0.163 | 0.1972 | No |
| 10 | Rtn1 | 5825 | 0.117 | 0.1621 | No |
| 11 | Rasa1 | 6151 | 0.101 | 0.1569 | No |
| 12 | Unc5c | 6773 | 0.071 | 0.1296 | No |
| 13 | Pml | 6815 | 0.069 | 0.1369 | No |
| 14 | Myh9 | 7790 | 0.025 | 0.0819 | No |
| 15 | Cdk6 | 9482 | -0.045 | -0.0131 | No |
| 16 | Dpysl2 | 9505 | -0.046 | -0.0078 | No |
| 17 | Nrp2 | 9665 | -0.053 | -0.0099 | No |
| 18 | Adgrg1 | 9877 | -0.064 | -0.0135 | No |
| 19 | Slit1 | 10006 | -0.069 | -0.0115 | No |
| 20 | Vldlr | 10252 | -0.080 | -0.0149 | No |
| 21 | Vegfa | 12373 | -0.180 | -0.1166 | No |
| 22 | Tle1 | 13019 | -0.215 | -0.1249 | No |
| 23 | L1cam | 13286 | -0.232 | -0.1080 | No |
| 24 | Thy1 | 13865 | -0.268 | -0.1048 | No |
| 25 | Tle3 | 14612 | -0.325 | -0.1037 | No |
| 26 | Plg | 15849 | -0.470 | -0.1113 | No |
| 27 | Ets2 | 16089 | -0.515 | -0.0529 | No |
| 28 | Cdk5r1 | 16435 | -0.633 | 0.0158 | No |