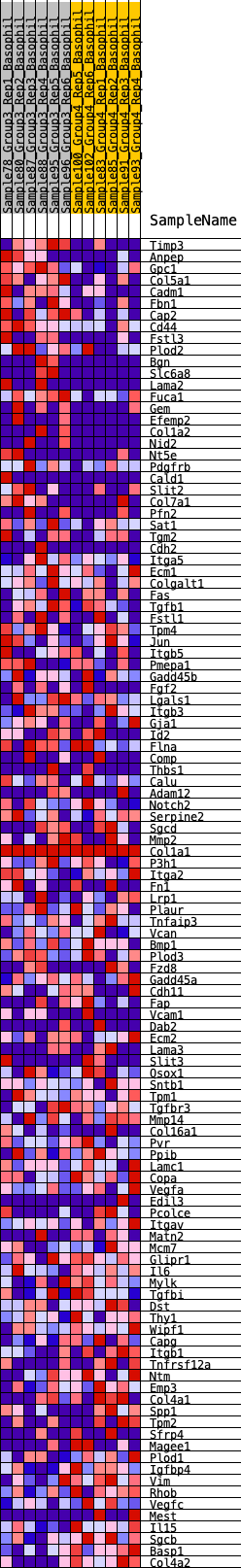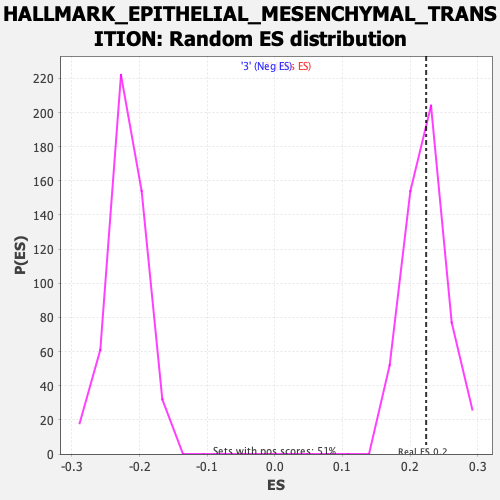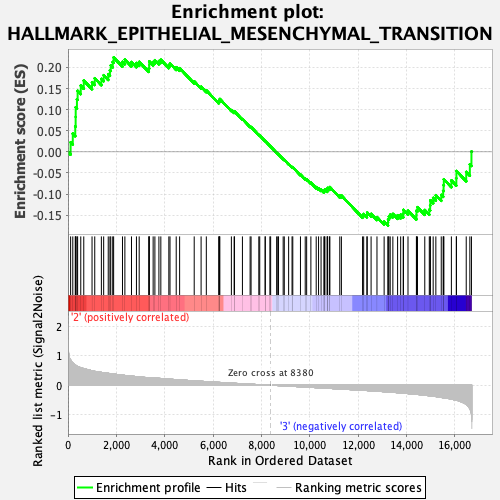
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.22380127 |
| Normalized Enrichment Score (NES) | 1.0022191 |
| Nominal p-value | 0.47758284 |
| FDR q-value | 0.8561005 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Timp3 | 108 | 0.859 | 0.0227 | Yes |
| 2 | Anpep | 198 | 0.762 | 0.0432 | Yes |
| 3 | Gpc1 | 300 | 0.688 | 0.0605 | Yes |
| 4 | Col5a1 | 316 | 0.682 | 0.0827 | Yes |
| 5 | Cadm1 | 319 | 0.678 | 0.1057 | Yes |
| 6 | Fbn1 | 370 | 0.648 | 0.1247 | Yes |
| 7 | Cap2 | 398 | 0.635 | 0.1446 | Yes |
| 8 | Cd44 | 528 | 0.595 | 0.1571 | Yes |
| 9 | Fstl3 | 652 | 0.568 | 0.1689 | Yes |
| 10 | Plod2 | 994 | 0.492 | 0.1651 | Yes |
| 11 | Bgn | 1106 | 0.472 | 0.1744 | Yes |
| 12 | Slc6a8 | 1380 | 0.437 | 0.1728 | Yes |
| 13 | Lama2 | 1475 | 0.426 | 0.1816 | Yes |
| 14 | Fuca1 | 1664 | 0.405 | 0.1841 | Yes |
| 15 | Gem | 1734 | 0.397 | 0.1934 | Yes |
| 16 | Efemp2 | 1768 | 0.395 | 0.2048 | Yes |
| 17 | Col1a2 | 1850 | 0.386 | 0.2130 | Yes |
| 18 | Nid2 | 1887 | 0.381 | 0.2238 | Yes |
| 19 | Nt5e | 2253 | 0.341 | 0.2134 | No |
| 20 | Pdgfrb | 2352 | 0.334 | 0.2188 | No |
| 21 | Cald1 | 2625 | 0.309 | 0.2129 | No |
| 22 | Slit2 | 2830 | 0.292 | 0.2105 | No |
| 23 | Col7a1 | 2944 | 0.283 | 0.2133 | No |
| 24 | Pfn2 | 3340 | 0.256 | 0.1982 | No |
| 25 | Sat1 | 3359 | 0.256 | 0.2058 | No |
| 26 | Tgm2 | 3360 | 0.255 | 0.2144 | No |
| 27 | Cdh2 | 3521 | 0.246 | 0.2132 | No |
| 28 | Itga5 | 3593 | 0.244 | 0.2172 | No |
| 29 | Ecm1 | 3759 | 0.235 | 0.2152 | No |
| 30 | Colgalt1 | 3839 | 0.230 | 0.2182 | No |
| 31 | Fas | 4170 | 0.207 | 0.2054 | No |
| 32 | Tgfb1 | 4218 | 0.203 | 0.2094 | No |
| 33 | Fstl1 | 4475 | 0.191 | 0.2005 | No |
| 34 | Tpm4 | 4616 | 0.183 | 0.1983 | No |
| 35 | Jun | 5219 | 0.149 | 0.1670 | No |
| 36 | Itgb5 | 5504 | 0.135 | 0.1545 | No |
| 37 | Pmepa1 | 5720 | 0.123 | 0.1457 | No |
| 38 | Gadd45b | 6234 | 0.097 | 0.1180 | No |
| 39 | Fgf2 | 6255 | 0.096 | 0.1201 | No |
| 40 | Lgals1 | 6260 | 0.096 | 0.1231 | No |
| 41 | Itgb3 | 6279 | 0.096 | 0.1253 | No |
| 42 | Gja1 | 6756 | 0.071 | 0.0990 | No |
| 43 | Id2 | 6870 | 0.067 | 0.0944 | No |
| 44 | Flna | 6886 | 0.066 | 0.0958 | No |
| 45 | Comp | 7213 | 0.051 | 0.0779 | No |
| 46 | Thbs1 | 7531 | 0.037 | 0.0600 | No |
| 47 | Calu | 7570 | 0.035 | 0.0589 | No |
| 48 | Adam12 | 7891 | 0.021 | 0.0403 | No |
| 49 | Notch2 | 7929 | 0.019 | 0.0387 | No |
| 50 | Serpine2 | 8152 | 0.010 | 0.0257 | No |
| 51 | Sgcd | 8158 | 0.009 | 0.0257 | No |
| 52 | Mmp2 | 8349 | 0.001 | 0.0143 | No |
| 53 | Col1a1 | 8380 | 0.000 | 0.0125 | No |
| 54 | P3h1 | 8628 | -0.008 | -0.0022 | No |
| 55 | Itga2 | 8675 | -0.010 | -0.0046 | No |
| 56 | Fn1 | 8706 | -0.011 | -0.0060 | No |
| 57 | Lrp1 | 8898 | -0.019 | -0.0169 | No |
| 58 | Plaur | 8944 | -0.021 | -0.0189 | No |
| 59 | Tnfaip3 | 9124 | -0.028 | -0.0287 | No |
| 60 | Vcan | 9265 | -0.035 | -0.0360 | No |
| 61 | Bmp1 | 9296 | -0.037 | -0.0365 | No |
| 62 | Plod3 | 9613 | -0.051 | -0.0539 | No |
| 63 | Fzd8 | 9810 | -0.060 | -0.0636 | No |
| 64 | Gadd45a | 9869 | -0.063 | -0.0650 | No |
| 65 | Cdh11 | 10044 | -0.071 | -0.0731 | No |
| 66 | Fap | 10264 | -0.081 | -0.0836 | No |
| 67 | Vcam1 | 10361 | -0.085 | -0.0864 | No |
| 68 | Dab2 | 10465 | -0.090 | -0.0896 | No |
| 69 | Ecm2 | 10582 | -0.096 | -0.0933 | No |
| 70 | Lama3 | 10605 | -0.097 | -0.0914 | No |
| 71 | Slit3 | 10624 | -0.098 | -0.0891 | No |
| 72 | Qsox1 | 10720 | -0.102 | -0.0914 | No |
| 73 | Sntb1 | 10722 | -0.102 | -0.0880 | No |
| 74 | Tpm1 | 10739 | -0.103 | -0.0854 | No |
| 75 | Tgfbr3 | 10818 | -0.106 | -0.0865 | No |
| 76 | Mmp14 | 10827 | -0.107 | -0.0834 | No |
| 77 | Col16a1 | 11239 | -0.126 | -0.1039 | No |
| 78 | Pvr | 11306 | -0.129 | -0.1035 | No |
| 79 | Ppib | 12181 | -0.170 | -0.1504 | No |
| 80 | Lamc1 | 12227 | -0.172 | -0.1472 | No |
| 81 | Copa | 12361 | -0.179 | -0.1492 | No |
| 82 | Vegfa | 12373 | -0.180 | -0.1437 | No |
| 83 | Edil3 | 12534 | -0.189 | -0.1469 | No |
| 84 | Pcolce | 12777 | -0.201 | -0.1547 | No |
| 85 | Itgav | 13073 | -0.219 | -0.1651 | No |
| 86 | Matn2 | 13234 | -0.228 | -0.1670 | No |
| 87 | Mcm7 | 13237 | -0.229 | -0.1593 | No |
| 88 | Glipr1 | 13271 | -0.231 | -0.1534 | No |
| 89 | Il6 | 13324 | -0.234 | -0.1486 | No |
| 90 | Mylk | 13434 | -0.241 | -0.1470 | No |
| 91 | Tgfbi | 13630 | -0.252 | -0.1502 | No |
| 92 | Dst | 13757 | -0.261 | -0.1489 | No |
| 93 | Thy1 | 13865 | -0.268 | -0.1463 | No |
| 94 | Wipf1 | 13870 | -0.268 | -0.1374 | No |
| 95 | Capg | 14060 | -0.283 | -0.1392 | No |
| 96 | Itgb1 | 14409 | -0.309 | -0.1497 | No |
| 97 | Tnfrsf12a | 14410 | -0.309 | -0.1392 | No |
| 98 | Ntm | 14452 | -0.312 | -0.1311 | No |
| 99 | Emp3 | 14754 | -0.337 | -0.1378 | No |
| 100 | Col4a1 | 14941 | -0.358 | -0.1369 | No |
| 101 | Spp1 | 14977 | -0.361 | -0.1267 | No |
| 102 | Tpm2 | 14984 | -0.362 | -0.1148 | No |
| 103 | Sfrp4 | 15101 | -0.373 | -0.1091 | No |
| 104 | Magee1 | 15212 | -0.383 | -0.1027 | No |
| 105 | Plod1 | 15435 | -0.414 | -0.1020 | No |
| 106 | Igfbp4 | 15514 | -0.424 | -0.0923 | No |
| 107 | Vim | 15532 | -0.426 | -0.0789 | No |
| 108 | Rhob | 15542 | -0.427 | -0.0649 | No |
| 109 | Vegfc | 15854 | -0.472 | -0.0677 | No |
| 110 | Mest | 16059 | -0.512 | -0.0626 | No |
| 111 | Il15 | 16063 | -0.512 | -0.0453 | No |
| 112 | Sgcb | 16468 | -0.653 | -0.0475 | No |
| 113 | Basp1 | 16618 | -0.795 | -0.0295 | No |
| 114 | Col4a2 | 16684 | -1.010 | 0.0009 | No |