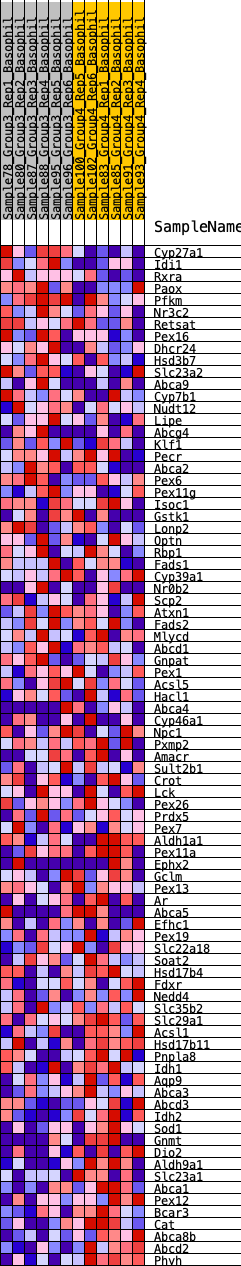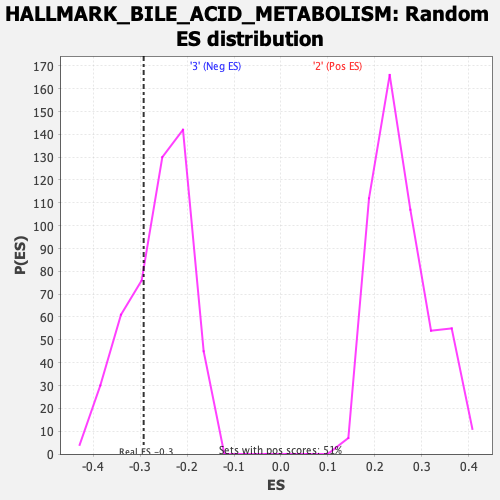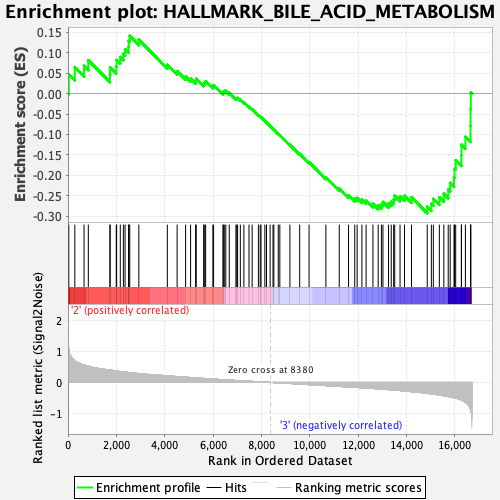
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.29258937 |
| Normalized Enrichment Score (NES) | -1.1274308 |
| Nominal p-value | 0.29508197 |
| FDR q-value | 0.7118582 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp27a1 | 33 | 1.067 | 0.0473 | No |
| 2 | Idi1 | 282 | 0.704 | 0.0649 | No |
| 3 | Rxra | 664 | 0.565 | 0.0681 | No |
| 4 | Paox | 840 | 0.526 | 0.0819 | No |
| 5 | Pfkm | 1735 | 0.397 | 0.0465 | No |
| 6 | Nr3c2 | 1742 | 0.396 | 0.0644 | No |
| 7 | Retsat | 1995 | 0.370 | 0.0663 | No |
| 8 | Pex16 | 2012 | 0.367 | 0.0823 | No |
| 9 | Dhcr24 | 2162 | 0.350 | 0.0895 | No |
| 10 | Hsd3b7 | 2290 | 0.338 | 0.0975 | No |
| 11 | Slc23a2 | 2360 | 0.333 | 0.1088 | No |
| 12 | Abca9 | 2503 | 0.320 | 0.1150 | No |
| 13 | Cyp7b1 | 2514 | 0.319 | 0.1291 | No |
| 14 | Nudt12 | 2553 | 0.315 | 0.1414 | No |
| 15 | Lipe | 2930 | 0.284 | 0.1319 | No |
| 16 | Abcg4 | 4108 | 0.211 | 0.0708 | No |
| 17 | Klf1 | 4513 | 0.189 | 0.0552 | No |
| 18 | Pecr | 4864 | 0.168 | 0.0419 | No |
| 19 | Abca2 | 5070 | 0.157 | 0.0368 | No |
| 20 | Pex6 | 5277 | 0.146 | 0.0311 | No |
| 21 | Pex11g | 5302 | 0.145 | 0.0364 | No |
| 22 | Isoc1 | 5608 | 0.129 | 0.0240 | No |
| 23 | Gstk1 | 5653 | 0.127 | 0.0272 | No |
| 24 | Lonp2 | 5692 | 0.125 | 0.0307 | No |
| 25 | Optn | 5991 | 0.109 | 0.0178 | No |
| 26 | Rbp1 | 6021 | 0.107 | 0.0210 | No |
| 27 | Fads1 | 6410 | 0.088 | 0.0017 | No |
| 28 | Cyp39a1 | 6422 | 0.088 | 0.0051 | No |
| 29 | Nr0b2 | 6463 | 0.086 | 0.0067 | No |
| 30 | Scp2 | 6526 | 0.082 | 0.0067 | No |
| 31 | Atxn1 | 6668 | 0.075 | 0.0017 | No |
| 32 | Fads2 | 6944 | 0.063 | -0.0120 | No |
| 33 | Mlycd | 6981 | 0.061 | -0.0113 | No |
| 34 | Abcd1 | 7014 | 0.060 | -0.0105 | No |
| 35 | Gnpat | 7127 | 0.055 | -0.0147 | No |
| 36 | Pex1 | 7273 | 0.048 | -0.0212 | No |
| 37 | Acsl5 | 7485 | 0.039 | -0.0321 | No |
| 38 | Hacl1 | 7615 | 0.033 | -0.0383 | No |
| 39 | Abca4 | 7873 | 0.022 | -0.0527 | No |
| 40 | Cyp46a1 | 7947 | 0.018 | -0.0563 | No |
| 41 | Npc1 | 7982 | 0.017 | -0.0576 | No |
| 42 | Pxmp2 | 8141 | 0.010 | -0.0666 | No |
| 43 | Amacr | 8209 | 0.007 | -0.0703 | No |
| 44 | Sult2b1 | 8358 | 0.001 | -0.0792 | No |
| 45 | Crot | 8478 | -0.001 | -0.0863 | No |
| 46 | Lck | 8512 | -0.002 | -0.0882 | No |
| 47 | Pex26 | 8696 | -0.011 | -0.0987 | No |
| 48 | Prdx5 | 8760 | -0.014 | -0.1018 | No |
| 49 | Pex7 | 9177 | -0.030 | -0.1255 | No |
| 50 | Aldh1a1 | 9581 | -0.049 | -0.1475 | No |
| 51 | Pex11a | 9969 | -0.068 | -0.1676 | No |
| 52 | Ephx2 | 10664 | -0.100 | -0.2048 | No |
| 53 | Gclm | 11217 | -0.125 | -0.2322 | No |
| 54 | Pex13 | 11601 | -0.143 | -0.2487 | No |
| 55 | Ar | 11858 | -0.155 | -0.2569 | No |
| 56 | Abca5 | 11957 | -0.159 | -0.2554 | No |
| 57 | Efhc1 | 12154 | -0.169 | -0.2594 | No |
| 58 | Pex19 | 12327 | -0.178 | -0.2616 | No |
| 59 | Slc22a18 | 12610 | -0.191 | -0.2697 | No |
| 60 | Soat2 | 12827 | -0.204 | -0.2733 | No |
| 61 | Hsd17b4 | 12961 | -0.212 | -0.2714 | No |
| 62 | Fdxr | 13024 | -0.216 | -0.2652 | No |
| 63 | Nedd4 | 13256 | -0.230 | -0.2685 | No |
| 64 | Slc35b2 | 13367 | -0.237 | -0.2641 | No |
| 65 | Slc29a1 | 13467 | -0.243 | -0.2589 | No |
| 66 | Acsl1 | 13511 | -0.246 | -0.2501 | No |
| 67 | Hsd17b11 | 13731 | -0.259 | -0.2513 | No |
| 68 | Pnpla8 | 13915 | -0.271 | -0.2498 | No |
| 69 | Idh1 | 14205 | -0.295 | -0.2535 | No |
| 70 | Aqp9 | 14855 | -0.348 | -0.2765 | Yes |
| 71 | Abca3 | 15026 | -0.366 | -0.2698 | Yes |
| 72 | Abcd3 | 15108 | -0.373 | -0.2574 | Yes |
| 73 | Idh2 | 15356 | -0.403 | -0.2537 | Yes |
| 74 | Sod1 | 15543 | -0.427 | -0.2451 | Yes |
| 75 | Gnmt | 15723 | -0.452 | -0.2350 | Yes |
| 76 | Dio2 | 15804 | -0.462 | -0.2185 | Yes |
| 77 | Aldh9a1 | 15964 | -0.492 | -0.2054 | Yes |
| 78 | Slc23a1 | 15985 | -0.496 | -0.1836 | Yes |
| 79 | Abca1 | 16032 | -0.507 | -0.1630 | Yes |
| 80 | Pex12 | 16268 | -0.564 | -0.1510 | Yes |
| 81 | Bcar3 | 16271 | -0.564 | -0.1251 | Yes |
| 82 | Cat | 16434 | -0.632 | -0.1056 | Yes |
| 83 | Abca8b | 16646 | -0.864 | -0.0784 | Yes |
| 84 | Abcd2 | 16649 | -0.868 | -0.0384 | Yes |
| 85 | Phyh | 16658 | -0.895 | 0.0025 | Yes |