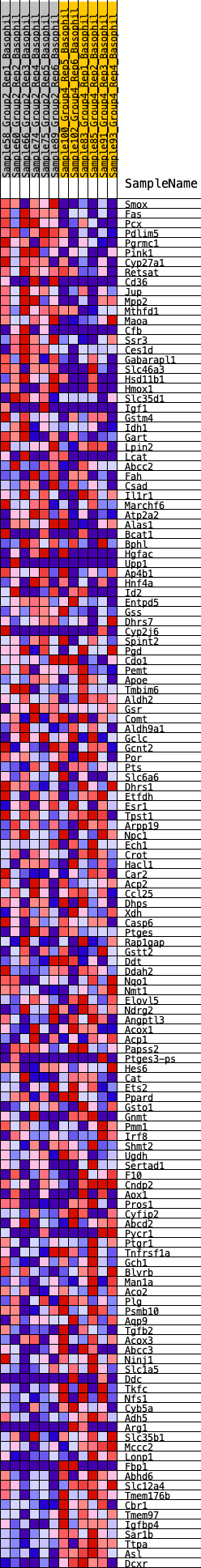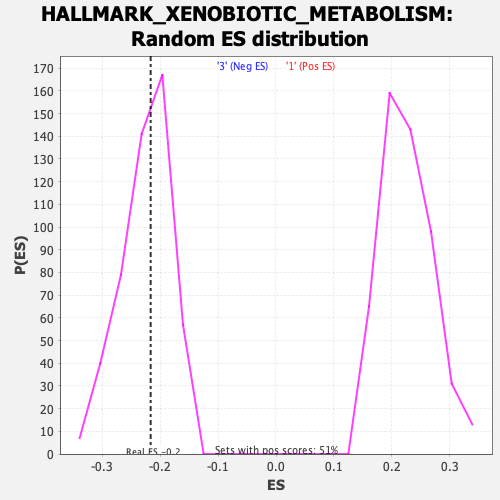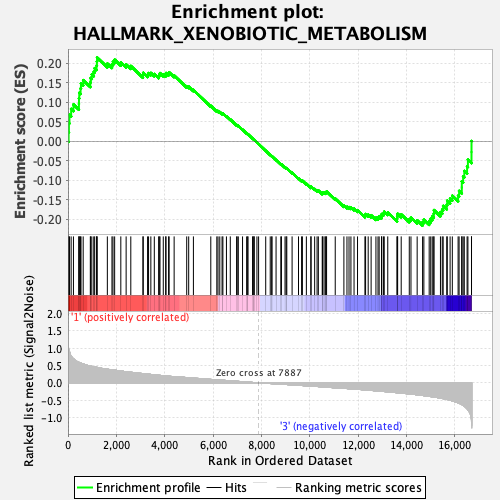
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.21617651 |
| Normalized Enrichment Score (NES) | -0.9593763 |
| Nominal p-value | 0.52953154 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smox | 26 | 1.000 | 0.0234 | Yes |
| 2 | Fas | 38 | 0.957 | 0.0466 | Yes |
| 3 | Pcx | 60 | 0.907 | 0.0680 | Yes |
| 4 | Pdlim5 | 130 | 0.767 | 0.0830 | Yes |
| 5 | Pgrmc1 | 225 | 0.692 | 0.0946 | Yes |
| 6 | Pink1 | 451 | 0.592 | 0.0958 | Yes |
| 7 | Cyp27a1 | 454 | 0.591 | 0.1104 | Yes |
| 8 | Retsat | 466 | 0.586 | 0.1244 | Yes |
| 9 | Cd36 | 520 | 0.570 | 0.1354 | Yes |
| 10 | Jup | 536 | 0.567 | 0.1487 | Yes |
| 11 | Mpp2 | 628 | 0.544 | 0.1568 | Yes |
| 12 | Mthfd1 | 923 | 0.484 | 0.1511 | Yes |
| 13 | Maoa | 937 | 0.482 | 0.1624 | Yes |
| 14 | Cfb | 985 | 0.475 | 0.1714 | Yes |
| 15 | Ssr3 | 1066 | 0.463 | 0.1781 | Yes |
| 16 | Ces1d | 1103 | 0.459 | 0.1874 | Yes |
| 17 | Gabarapl1 | 1182 | 0.449 | 0.1939 | Yes |
| 18 | Slc46a3 | 1194 | 0.448 | 0.2044 | Yes |
| 19 | Hsd11b1 | 1203 | 0.446 | 0.2151 | Yes |
| 20 | Hmox1 | 1627 | 0.393 | 0.1994 | No |
| 21 | Slc35d1 | 1818 | 0.376 | 0.1973 | No |
| 22 | Igf1 | 1860 | 0.373 | 0.2041 | No |
| 23 | Gstm4 | 1929 | 0.368 | 0.2092 | No |
| 24 | Idh1 | 2184 | 0.341 | 0.2023 | No |
| 25 | Gart | 2404 | 0.322 | 0.1972 | No |
| 26 | Lpin2 | 2597 | 0.305 | 0.1932 | No |
| 27 | Lcat | 3099 | 0.266 | 0.1696 | No |
| 28 | Abcc2 | 3110 | 0.266 | 0.1756 | No |
| 29 | Fah | 3291 | 0.252 | 0.1710 | No |
| 30 | Csad | 3331 | 0.250 | 0.1749 | No |
| 31 | Il1r1 | 3426 | 0.243 | 0.1753 | No |
| 32 | Marchf6 | 3569 | 0.232 | 0.1725 | No |
| 33 | Atp2a2 | 3749 | 0.219 | 0.1672 | No |
| 34 | Alas1 | 3774 | 0.217 | 0.1712 | No |
| 35 | Bcat1 | 3804 | 0.215 | 0.1748 | No |
| 36 | Bphl | 3940 | 0.207 | 0.1718 | No |
| 37 | Hgfac | 4040 | 0.199 | 0.1708 | No |
| 38 | Upp1 | 4052 | 0.198 | 0.1751 | No |
| 39 | Ap4b1 | 4160 | 0.192 | 0.1734 | No |
| 40 | Hnf4a | 4181 | 0.191 | 0.1770 | No |
| 41 | Id2 | 4389 | 0.178 | 0.1689 | No |
| 42 | Entpd5 | 4909 | 0.152 | 0.1414 | No |
| 43 | Gss | 4991 | 0.148 | 0.1402 | No |
| 44 | Dhrs7 | 5186 | 0.138 | 0.1319 | No |
| 45 | Cyp2j6 | 5905 | 0.100 | 0.0910 | No |
| 46 | Spint2 | 6154 | 0.088 | 0.0782 | No |
| 47 | Pgd | 6203 | 0.085 | 0.0775 | No |
| 48 | Cdo1 | 6277 | 0.082 | 0.0751 | No |
| 49 | Pemt | 6371 | 0.077 | 0.0714 | No |
| 50 | Apoe | 6405 | 0.075 | 0.0713 | No |
| 51 | Tmbim6 | 6552 | 0.068 | 0.0642 | No |
| 52 | Aldh2 | 6711 | 0.060 | 0.0561 | No |
| 53 | Gsr | 6972 | 0.046 | 0.0416 | No |
| 54 | Comt | 6996 | 0.045 | 0.0413 | No |
| 55 | Aldh9a1 | 7043 | 0.043 | 0.0396 | No |
| 56 | Gclc | 7218 | 0.033 | 0.0299 | No |
| 57 | Gcnt2 | 7387 | 0.024 | 0.0204 | No |
| 58 | Por | 7431 | 0.022 | 0.0183 | No |
| 59 | Pts | 7441 | 0.021 | 0.0183 | No |
| 60 | Slc6a6 | 7632 | 0.013 | 0.0072 | No |
| 61 | Dhrs1 | 7657 | 0.011 | 0.0060 | No |
| 62 | Etfdh | 7689 | 0.010 | 0.0044 | No |
| 63 | Esr1 | 7701 | 0.009 | 0.0039 | No |
| 64 | Tpst1 | 7813 | 0.004 | -0.0027 | No |
| 65 | Arpp19 | 7882 | 0.000 | -0.0068 | No |
| 66 | Npc1 | 8176 | -0.010 | -0.0242 | No |
| 67 | Ech1 | 8363 | -0.017 | -0.0350 | No |
| 68 | Crot | 8424 | -0.020 | -0.0382 | No |
| 69 | Hacl1 | 8436 | -0.020 | -0.0383 | No |
| 70 | Car2 | 8604 | -0.028 | -0.0477 | No |
| 71 | Acp2 | 8801 | -0.037 | -0.0586 | No |
| 72 | Ccl25 | 8827 | -0.038 | -0.0592 | No |
| 73 | Dhps | 8975 | -0.043 | -0.0670 | No |
| 74 | Xdh | 9022 | -0.045 | -0.0686 | No |
| 75 | Casp6 | 9048 | -0.046 | -0.0690 | No |
| 76 | Ptges | 9267 | -0.055 | -0.0808 | No |
| 77 | Rap1gap | 9532 | -0.068 | -0.0950 | No |
| 78 | Gstt2 | 9660 | -0.074 | -0.1008 | No |
| 79 | Ddt | 9693 | -0.075 | -0.1009 | No |
| 80 | Ddah2 | 9857 | -0.082 | -0.1087 | No |
| 81 | Nqo1 | 10042 | -0.091 | -0.1175 | No |
| 82 | Nmt1 | 10052 | -0.091 | -0.1158 | No |
| 83 | Elovl5 | 10200 | -0.097 | -0.1222 | No |
| 84 | Ndrg2 | 10300 | -0.102 | -0.1257 | No |
| 85 | Angptl3 | 10353 | -0.105 | -0.1262 | No |
| 86 | Acox1 | 10513 | -0.114 | -0.1329 | No |
| 87 | Acp1 | 10527 | -0.114 | -0.1309 | No |
| 88 | Papss2 | 10587 | -0.117 | -0.1315 | No |
| 89 | Ptges3-ps | 10642 | -0.120 | -0.1318 | No |
| 90 | Hes6 | 10670 | -0.121 | -0.1304 | No |
| 91 | Cat | 10693 | -0.122 | -0.1287 | No |
| 92 | Ets2 | 11051 | -0.141 | -0.1467 | No |
| 93 | Ppard | 11409 | -0.156 | -0.1644 | No |
| 94 | Gsto1 | 11536 | -0.162 | -0.1680 | No |
| 95 | Gnmt | 11621 | -0.167 | -0.1688 | No |
| 96 | Pmm1 | 11700 | -0.171 | -0.1693 | No |
| 97 | Irf8 | 11829 | -0.177 | -0.1726 | No |
| 98 | Shmt2 | 11973 | -0.185 | -0.1766 | No |
| 99 | Ugdh | 12279 | -0.201 | -0.1900 | No |
| 100 | Sertad1 | 12306 | -0.202 | -0.1865 | No |
| 101 | F10 | 12414 | -0.208 | -0.1878 | No |
| 102 | Cndp2 | 12535 | -0.215 | -0.1897 | No |
| 103 | Aox1 | 12734 | -0.228 | -0.1960 | No |
| 104 | Pros1 | 12820 | -0.232 | -0.1953 | No |
| 105 | Cyfip2 | 12867 | -0.235 | -0.1922 | No |
| 106 | Abcd2 | 12964 | -0.241 | -0.1920 | No |
| 107 | Pycr1 | 12975 | -0.242 | -0.1866 | No |
| 108 | Ptgr1 | 13044 | -0.246 | -0.1845 | No |
| 109 | Tnfrsf1a | 13077 | -0.248 | -0.1803 | No |
| 110 | Gch1 | 13223 | -0.256 | -0.1826 | No |
| 111 | Blvrb | 13608 | -0.282 | -0.1988 | No |
| 112 | Man1a | 13613 | -0.283 | -0.1919 | No |
| 113 | Aco2 | 13632 | -0.284 | -0.1859 | No |
| 114 | Plg | 13779 | -0.294 | -0.1874 | No |
| 115 | Psmb10 | 14113 | -0.314 | -0.1997 | No |
| 116 | Aqp9 | 14183 | -0.320 | -0.1959 | No |
| 117 | Tgfb2 | 14439 | -0.342 | -0.2027 | No |
| 118 | Acox3 | 14663 | -0.362 | -0.2071 | No |
| 119 | Abcc3 | 14716 | -0.366 | -0.2011 | No |
| 120 | Ninj1 | 14936 | -0.386 | -0.2047 | No |
| 121 | Slc1a5 | 15000 | -0.393 | -0.1987 | No |
| 122 | Ddc | 15063 | -0.400 | -0.1925 | No |
| 123 | Tkfc | 15125 | -0.405 | -0.1861 | No |
| 124 | Nfs1 | 15134 | -0.406 | -0.1764 | No |
| 125 | Cyb5a | 15402 | -0.439 | -0.1816 | No |
| 126 | Adh5 | 15476 | -0.448 | -0.1748 | No |
| 127 | Arg1 | 15514 | -0.452 | -0.1657 | No |
| 128 | Slc35b1 | 15664 | -0.472 | -0.1629 | No |
| 129 | Mccc2 | 15683 | -0.476 | -0.1521 | No |
| 130 | Lonp1 | 15801 | -0.497 | -0.1468 | No |
| 131 | Fbp1 | 15893 | -0.516 | -0.1394 | No |
| 132 | Abhd6 | 16132 | -0.575 | -0.1394 | No |
| 133 | Slc12a4 | 16173 | -0.587 | -0.1272 | No |
| 134 | Tmem176b | 16285 | -0.621 | -0.1184 | No |
| 135 | Cbr1 | 16286 | -0.622 | -0.1028 | No |
| 136 | Tmem97 | 16340 | -0.647 | -0.0899 | No |
| 137 | Igfbp4 | 16388 | -0.666 | -0.0761 | No |
| 138 | Sar1b | 16504 | -0.745 | -0.0644 | No |
| 139 | Ttpa | 16538 | -0.774 | -0.0471 | No |
| 140 | Asl | 16686 | -1.134 | -0.0277 | No |
| 141 | Dcxr | 16687 | -1.137 | 0.0007 | No |