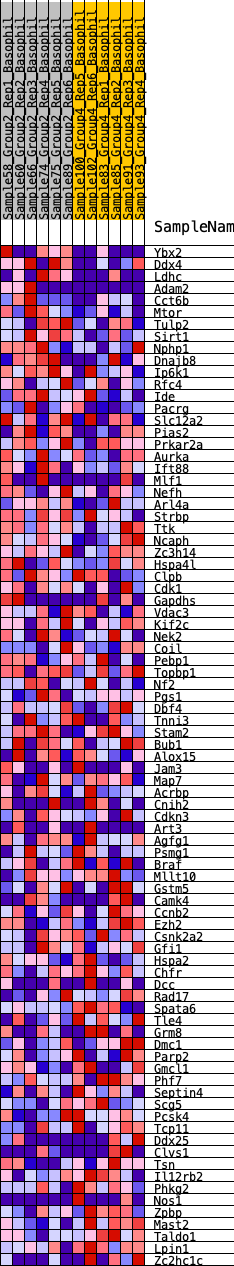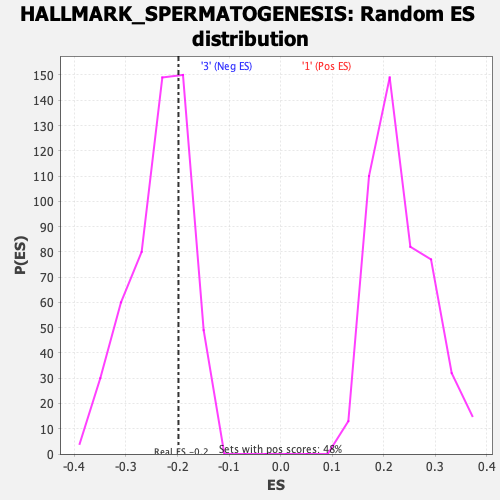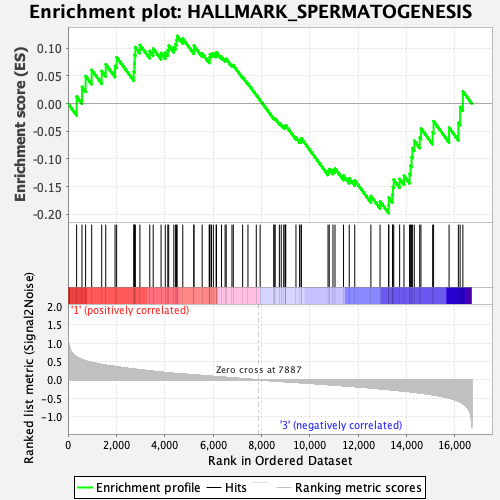
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.1981262 |
| Normalized Enrichment Score (NES) | -0.84750545 |
| Nominal p-value | 0.697318 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ybx2 | 356 | 0.624 | 0.0129 | No |
| 2 | Ddx4 | 578 | 0.558 | 0.0302 | No |
| 3 | Ldhc | 728 | 0.521 | 0.0499 | No |
| 4 | Adam2 | 979 | 0.476 | 0.0610 | No |
| 5 | Cct6b | 1394 | 0.418 | 0.0590 | No |
| 6 | Mtor | 1561 | 0.400 | 0.0710 | No |
| 7 | Tulp2 | 1946 | 0.367 | 0.0681 | No |
| 8 | Sirt1 | 2011 | 0.359 | 0.0839 | No |
| 9 | Nphp1 | 2718 | 0.297 | 0.0577 | No |
| 10 | Dnajb8 | 2744 | 0.295 | 0.0724 | No |
| 11 | Ip6k1 | 2757 | 0.294 | 0.0878 | No |
| 12 | Rfc4 | 2785 | 0.291 | 0.1022 | No |
| 13 | Ide | 2973 | 0.275 | 0.1061 | No |
| 14 | Pacrg | 3381 | 0.246 | 0.0951 | No |
| 15 | Slc12a2 | 3523 | 0.235 | 0.0996 | No |
| 16 | Pias2 | 3849 | 0.213 | 0.0917 | No |
| 17 | Prkar2a | 4023 | 0.200 | 0.0922 | No |
| 18 | Aurka | 4127 | 0.195 | 0.0967 | No |
| 19 | Ift88 | 4164 | 0.192 | 0.1051 | No |
| 20 | Mlf1 | 4376 | 0.178 | 0.1022 | No |
| 21 | Nefh | 4449 | 0.174 | 0.1074 | No |
| 22 | Arl4a | 4485 | 0.172 | 0.1148 | No |
| 23 | Strbp | 4514 | 0.170 | 0.1224 | No |
| 24 | Ttk | 4746 | 0.162 | 0.1174 | No |
| 25 | Ncaph | 5205 | 0.136 | 0.0974 | No |
| 26 | Zc3h14 | 5207 | 0.136 | 0.1048 | No |
| 27 | Hspa4l | 5548 | 0.119 | 0.0909 | No |
| 28 | Clpb | 5839 | 0.103 | 0.0791 | No |
| 29 | Cdk1 | 5857 | 0.102 | 0.0837 | No |
| 30 | Gapdhs | 5870 | 0.101 | 0.0885 | No |
| 31 | Vdac3 | 5935 | 0.098 | 0.0901 | No |
| 32 | Kif2c | 6012 | 0.095 | 0.0907 | No |
| 33 | Nek2 | 6125 | 0.090 | 0.0889 | No |
| 34 | Coil | 6142 | 0.089 | 0.0928 | No |
| 35 | Pebp1 | 6349 | 0.078 | 0.0847 | No |
| 36 | Topbp1 | 6497 | 0.071 | 0.0797 | No |
| 37 | Nf2 | 6542 | 0.069 | 0.0809 | No |
| 38 | Pgs1 | 6780 | 0.056 | 0.0697 | No |
| 39 | Dbf4 | 6836 | 0.053 | 0.0692 | No |
| 40 | Tnni3 | 7222 | 0.033 | 0.0479 | No |
| 41 | Stam2 | 7442 | 0.021 | 0.0359 | No |
| 42 | Bub1 | 7786 | 0.005 | 0.0155 | No |
| 43 | Alox15 | 7946 | -0.000 | 0.0060 | No |
| 44 | Jam3 | 8506 | -0.024 | -0.0264 | No |
| 45 | Map7 | 8556 | -0.025 | -0.0279 | No |
| 46 | Acrbp | 8560 | -0.026 | -0.0267 | No |
| 47 | Cnih2 | 8741 | -0.034 | -0.0357 | No |
| 48 | Cdkn3 | 8818 | -0.038 | -0.0382 | No |
| 49 | Art3 | 8920 | -0.042 | -0.0419 | No |
| 50 | Agfg1 | 8977 | -0.043 | -0.0429 | No |
| 51 | Psmg1 | 8988 | -0.044 | -0.0411 | No |
| 52 | Braf | 9004 | -0.044 | -0.0396 | No |
| 53 | Mllt10 | 9427 | -0.063 | -0.0615 | No |
| 54 | Gstm5 | 9576 | -0.070 | -0.0666 | No |
| 55 | Camk4 | 9640 | -0.073 | -0.0664 | No |
| 56 | Ccnb2 | 9650 | -0.073 | -0.0629 | No |
| 57 | Ezh2 | 10753 | -0.125 | -0.1224 | No |
| 58 | Csnk2a2 | 10803 | -0.128 | -0.1183 | No |
| 59 | Gfi1 | 10957 | -0.136 | -0.1200 | No |
| 60 | Hspa2 | 11041 | -0.140 | -0.1173 | No |
| 61 | Chfr | 11394 | -0.154 | -0.1300 | No |
| 62 | Dcc | 11630 | -0.168 | -0.1349 | No |
| 63 | Rad17 | 11858 | -0.178 | -0.1388 | No |
| 64 | Spata6 | 12526 | -0.215 | -0.1671 | No |
| 65 | Tle4 | 12904 | -0.236 | -0.1768 | No |
| 66 | Grm8 | 13259 | -0.259 | -0.1839 | Yes |
| 67 | Dmc1 | 13267 | -0.259 | -0.1701 | Yes |
| 68 | Parp2 | 13418 | -0.269 | -0.1643 | Yes |
| 69 | Gmcl1 | 13437 | -0.270 | -0.1506 | Yes |
| 70 | Phf7 | 13471 | -0.273 | -0.1376 | Yes |
| 71 | Septin4 | 13714 | -0.289 | -0.1362 | Yes |
| 72 | Scg5 | 13894 | -0.301 | -0.1305 | Yes |
| 73 | Pcsk4 | 14124 | -0.315 | -0.1269 | Yes |
| 74 | Tcp11 | 14167 | -0.319 | -0.1119 | Yes |
| 75 | Ddx25 | 14209 | -0.322 | -0.0967 | Yes |
| 76 | Clvs1 | 14242 | -0.325 | -0.0808 | Yes |
| 77 | Tsn | 14319 | -0.332 | -0.0671 | Yes |
| 78 | Il12rb2 | 14545 | -0.352 | -0.0613 | Yes |
| 79 | Phkg2 | 14596 | -0.356 | -0.0448 | Yes |
| 80 | Nos1 | 15080 | -0.400 | -0.0518 | Yes |
| 81 | Zpbp | 15118 | -0.404 | -0.0318 | Yes |
| 82 | Mast2 | 15759 | -0.489 | -0.0435 | Yes |
| 83 | Taldo1 | 16143 | -0.578 | -0.0348 | Yes |
| 84 | Lpin1 | 16215 | -0.598 | -0.0062 | Yes |
| 85 | Zc2hc1c | 16329 | -0.641 | 0.0223 | Yes |