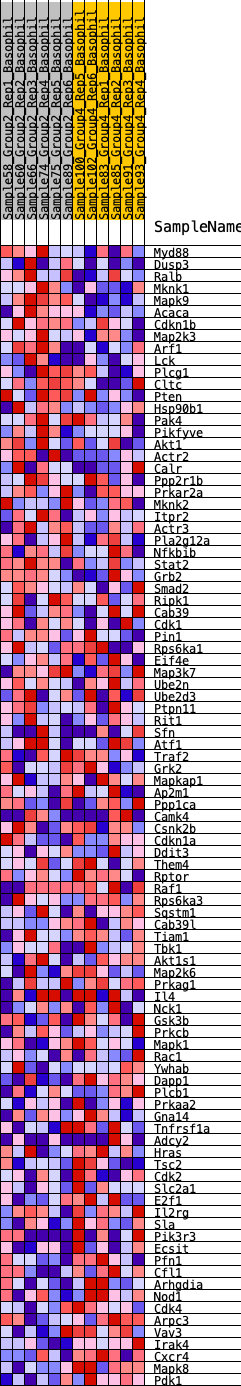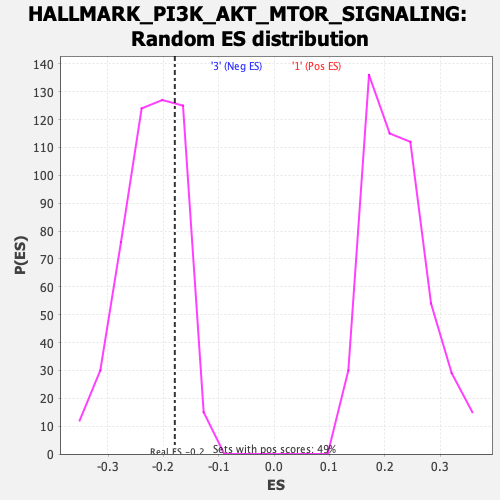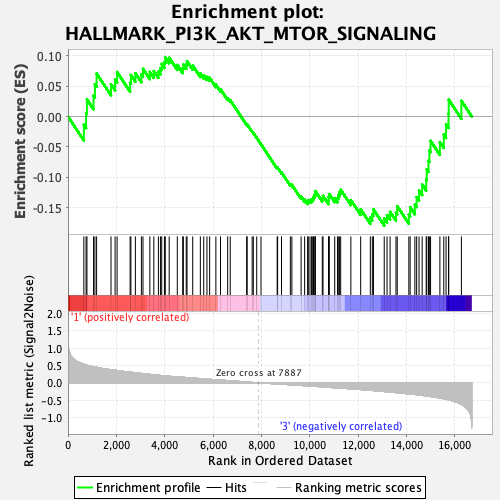
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.17929453 |
| Normalized Enrichment Score (NES) | -0.8098774 |
| Nominal p-value | 0.7721022 |
| FDR q-value | 0.98043567 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myd88 | 656 | 0.537 | -0.0135 | No |
| 2 | Dusp3 | 748 | 0.515 | 0.0059 | No |
| 3 | Ralb | 783 | 0.506 | 0.0283 | No |
| 4 | Mknk1 | 1056 | 0.465 | 0.0344 | No |
| 5 | Mapk9 | 1113 | 0.457 | 0.0531 | No |
| 6 | Acaca | 1183 | 0.449 | 0.0707 | No |
| 7 | Cdkn1b | 1778 | 0.379 | 0.0532 | No |
| 8 | Map2k3 | 1947 | 0.367 | 0.0608 | No |
| 9 | Arf1 | 2030 | 0.356 | 0.0731 | No |
| 10 | Lck | 2568 | 0.307 | 0.0556 | No |
| 11 | Plcg1 | 2601 | 0.304 | 0.0684 | No |
| 12 | Cltc | 2786 | 0.291 | 0.0714 | No |
| 13 | Pten | 3035 | 0.271 | 0.0696 | No |
| 14 | Hsp90b1 | 3101 | 0.266 | 0.0785 | No |
| 15 | Pak4 | 3384 | 0.246 | 0.0734 | No |
| 16 | Pikfyve | 3549 | 0.234 | 0.0748 | No |
| 17 | Akt1 | 3738 | 0.220 | 0.0741 | No |
| 18 | Actr2 | 3823 | 0.215 | 0.0795 | No |
| 19 | Calr | 3876 | 0.211 | 0.0865 | No |
| 20 | Ppp2r1b | 3988 | 0.203 | 0.0896 | No |
| 21 | Prkar2a | 4023 | 0.200 | 0.0972 | No |
| 22 | Mknk2 | 4184 | 0.190 | 0.0968 | No |
| 23 | Itpr2 | 4520 | 0.170 | 0.0848 | No |
| 24 | Actr3 | 4744 | 0.162 | 0.0792 | No |
| 25 | Pla2g12a | 4770 | 0.161 | 0.0855 | No |
| 26 | Nfkbib | 4890 | 0.154 | 0.0858 | No |
| 27 | Stat2 | 4922 | 0.152 | 0.0912 | No |
| 28 | Grb2 | 5157 | 0.139 | 0.0839 | No |
| 29 | Smad2 | 5473 | 0.124 | 0.0709 | No |
| 30 | Ripk1 | 5613 | 0.115 | 0.0681 | No |
| 31 | Cab39 | 5746 | 0.108 | 0.0654 | No |
| 32 | Cdk1 | 5857 | 0.102 | 0.0637 | No |
| 33 | Pin1 | 6115 | 0.090 | 0.0525 | No |
| 34 | Rps6ka1 | 6307 | 0.079 | 0.0449 | No |
| 35 | Eif4e | 6603 | 0.066 | 0.0303 | No |
| 36 | Map3k7 | 6709 | 0.060 | 0.0269 | No |
| 37 | Ube2n | 7388 | 0.024 | -0.0128 | No |
| 38 | Ube2d3 | 7408 | 0.023 | -0.0128 | No |
| 39 | Ptpn11 | 7616 | 0.013 | -0.0246 | No |
| 40 | Rit1 | 7663 | 0.011 | -0.0269 | No |
| 41 | Sfn | 7804 | 0.004 | -0.0351 | No |
| 42 | Atf1 | 7982 | -0.002 | -0.0457 | No |
| 43 | Traf2 | 8650 | -0.030 | -0.0844 | No |
| 44 | Grk2 | 8669 | -0.030 | -0.0841 | No |
| 45 | Mapkap1 | 8829 | -0.039 | -0.0918 | No |
| 46 | Ap2m1 | 9195 | -0.052 | -0.1112 | No |
| 47 | Ppp1ca | 9258 | -0.055 | -0.1123 | No |
| 48 | Camk4 | 9640 | -0.073 | -0.1317 | No |
| 49 | Csnk2b | 9786 | -0.080 | -0.1366 | No |
| 50 | Cdkn1a | 9910 | -0.084 | -0.1399 | No |
| 51 | Ddit3 | 9936 | -0.086 | -0.1373 | No |
| 52 | Them4 | 10007 | -0.089 | -0.1372 | No |
| 53 | Rptor | 10073 | -0.092 | -0.1367 | No |
| 54 | Raf1 | 10120 | -0.094 | -0.1349 | No |
| 55 | Rps6ka3 | 10152 | -0.095 | -0.1322 | No |
| 56 | Sqstm1 | 10181 | -0.097 | -0.1292 | No |
| 57 | Cab39l | 10220 | -0.098 | -0.1267 | No |
| 58 | Tiam1 | 10229 | -0.099 | -0.1224 | No |
| 59 | Tbk1 | 10515 | -0.114 | -0.1341 | No |
| 60 | Akt1s1 | 10546 | -0.115 | -0.1304 | No |
| 61 | Map2k6 | 10773 | -0.126 | -0.1379 | No |
| 62 | Prkag1 | 10781 | -0.126 | -0.1322 | No |
| 63 | Il4 | 10805 | -0.128 | -0.1274 | No |
| 64 | Nck1 | 11030 | -0.140 | -0.1341 | No |
| 65 | Gsk3b | 11146 | -0.144 | -0.1340 | No |
| 66 | Prkcb | 11176 | -0.146 | -0.1287 | No |
| 67 | Mapk1 | 11223 | -0.148 | -0.1243 | No |
| 68 | Rac1 | 11276 | -0.150 | -0.1202 | No |
| 69 | Ywhab | 11697 | -0.171 | -0.1373 | No |
| 70 | Dapp1 | 12103 | -0.191 | -0.1525 | No |
| 71 | Plcb1 | 12502 | -0.214 | -0.1661 | No |
| 72 | Prkaa2 | 12585 | -0.218 | -0.1605 | No |
| 73 | Gna14 | 12632 | -0.222 | -0.1526 | No |
| 74 | Tnfrsf1a | 13077 | -0.248 | -0.1673 | Yes |
| 75 | Adcy2 | 13194 | -0.255 | -0.1620 | Yes |
| 76 | Hras | 13319 | -0.262 | -0.1567 | Yes |
| 77 | Tsc2 | 13563 | -0.279 | -0.1579 | Yes |
| 78 | Cdk2 | 13615 | -0.283 | -0.1473 | Yes |
| 79 | Slc2a1 | 14095 | -0.313 | -0.1610 | Yes |
| 80 | E2f1 | 14148 | -0.317 | -0.1488 | Yes |
| 81 | Il2rg | 14345 | -0.334 | -0.1445 | Yes |
| 82 | Sla | 14418 | -0.340 | -0.1324 | Yes |
| 83 | Pik3r3 | 14516 | -0.349 | -0.1214 | Yes |
| 84 | Ecsit | 14646 | -0.361 | -0.1117 | Yes |
| 85 | Pfn1 | 14812 | -0.375 | -0.1035 | Yes |
| 86 | Cfl1 | 14836 | -0.376 | -0.0868 | Yes |
| 87 | Arhgdia | 14907 | -0.383 | -0.0725 | Yes |
| 88 | Nod1 | 14944 | -0.387 | -0.0559 | Yes |
| 89 | Cdk4 | 14993 | -0.392 | -0.0399 | Yes |
| 90 | Arpc3 | 15379 | -0.436 | -0.0420 | Yes |
| 91 | Vav3 | 15540 | -0.454 | -0.0297 | Yes |
| 92 | Irak4 | 15634 | -0.467 | -0.0127 | Yes |
| 93 | Cxcr4 | 15737 | -0.486 | 0.0047 | Yes |
| 94 | Mapk8 | 15744 | -0.487 | 0.0278 | Yes |
| 95 | Pdk1 | 16268 | -0.613 | 0.0260 | Yes |