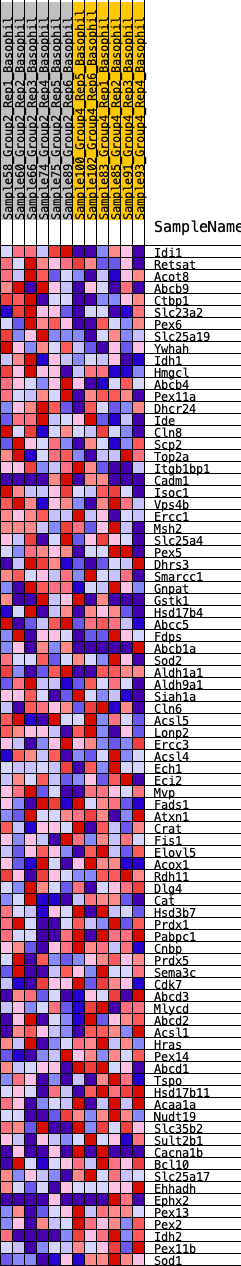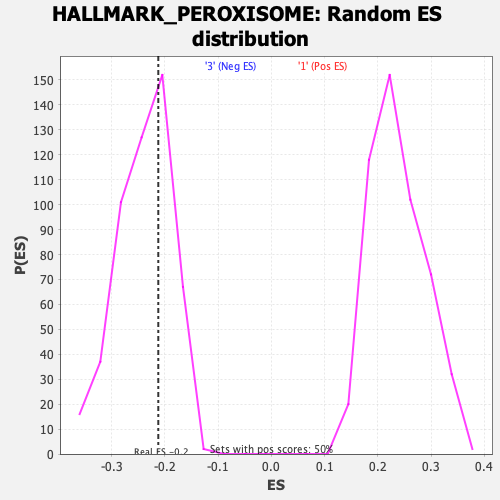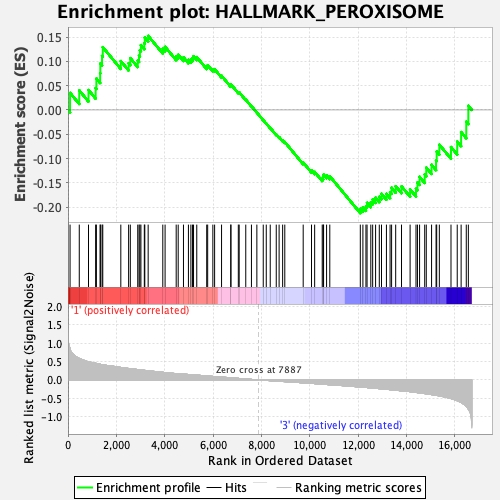
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.21226303 |
| Normalized Enrichment Score (NES) | -0.8915037 |
| Nominal p-value | 0.64143425 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 85 | 0.842 | 0.0350 | No |
| 2 | Retsat | 466 | 0.586 | 0.0401 | No |
| 3 | Acot8 | 848 | 0.497 | 0.0409 | No |
| 4 | Abcb9 | 1143 | 0.454 | 0.0449 | No |
| 5 | Ctbp1 | 1174 | 0.450 | 0.0646 | No |
| 6 | Slc23a2 | 1330 | 0.427 | 0.0756 | No |
| 7 | Pex6 | 1337 | 0.426 | 0.0956 | No |
| 8 | Slc25a19 | 1408 | 0.416 | 0.1112 | No |
| 9 | Ywhah | 1438 | 0.413 | 0.1291 | No |
| 10 | Idh1 | 2184 | 0.341 | 0.1006 | No |
| 11 | Hmgcl | 2507 | 0.313 | 0.0961 | No |
| 12 | Abcb4 | 2577 | 0.306 | 0.1066 | No |
| 13 | Pex11a | 2884 | 0.283 | 0.1016 | No |
| 14 | Dhcr24 | 2938 | 0.278 | 0.1117 | No |
| 15 | Ide | 2973 | 0.275 | 0.1228 | No |
| 16 | Cln8 | 3015 | 0.273 | 0.1334 | No |
| 17 | Scp2 | 3156 | 0.262 | 0.1374 | No |
| 18 | Top2a | 3176 | 0.260 | 0.1487 | No |
| 19 | Itgb1bp1 | 3315 | 0.251 | 0.1523 | No |
| 20 | Cadm1 | 3920 | 0.208 | 0.1259 | No |
| 21 | Isoc1 | 4015 | 0.201 | 0.1298 | No |
| 22 | Vps4b | 4474 | 0.173 | 0.1105 | No |
| 23 | Ercc1 | 4557 | 0.169 | 0.1136 | No |
| 24 | Msh2 | 4775 | 0.160 | 0.1082 | No |
| 25 | Slc25a4 | 4980 | 0.149 | 0.1030 | No |
| 26 | Pex5 | 5081 | 0.143 | 0.1038 | No |
| 27 | Dhrs3 | 5147 | 0.140 | 0.1066 | No |
| 28 | Smarcc1 | 5191 | 0.137 | 0.1105 | No |
| 29 | Gnpat | 5325 | 0.129 | 0.1087 | No |
| 30 | Gstk1 | 5736 | 0.109 | 0.0892 | No |
| 31 | Hsd17b4 | 5776 | 0.107 | 0.0920 | No |
| 32 | Abcc5 | 5990 | 0.096 | 0.0837 | No |
| 33 | Fdps | 6063 | 0.093 | 0.0838 | No |
| 34 | Abcb1a | 6350 | 0.078 | 0.0703 | No |
| 35 | Sod2 | 6731 | 0.059 | 0.0502 | No |
| 36 | Aldh1a1 | 6738 | 0.058 | 0.0527 | No |
| 37 | Aldh9a1 | 7043 | 0.043 | 0.0364 | No |
| 38 | Siah1a | 7079 | 0.041 | 0.0363 | No |
| 39 | Cln6 | 7351 | 0.026 | 0.0212 | No |
| 40 | Acsl5 | 7586 | 0.015 | 0.0079 | No |
| 41 | Lonp2 | 7810 | 0.004 | -0.0054 | No |
| 42 | Ercc3 | 8074 | -0.005 | -0.0210 | No |
| 43 | Acsl4 | 8199 | -0.011 | -0.0279 | No |
| 44 | Ech1 | 8363 | -0.017 | -0.0369 | No |
| 45 | Eci2 | 8612 | -0.028 | -0.0505 | No |
| 46 | Mvp | 8730 | -0.034 | -0.0559 | No |
| 47 | Fads1 | 8876 | -0.040 | -0.0627 | No |
| 48 | Atxn1 | 8965 | -0.043 | -0.0660 | No |
| 49 | Crat | 9726 | -0.077 | -0.1081 | No |
| 50 | Fis1 | 10070 | -0.092 | -0.1243 | No |
| 51 | Elovl5 | 10200 | -0.097 | -0.1275 | No |
| 52 | Acox1 | 10513 | -0.114 | -0.1408 | No |
| 53 | Rdh11 | 10553 | -0.116 | -0.1377 | No |
| 54 | Dlg4 | 10569 | -0.116 | -0.1330 | No |
| 55 | Cat | 10693 | -0.122 | -0.1346 | No |
| 56 | Hsd3b7 | 10826 | -0.129 | -0.1364 | No |
| 57 | Prdx1 | 12088 | -0.190 | -0.2032 | Yes |
| 58 | Pabpc1 | 12195 | -0.197 | -0.2002 | Yes |
| 59 | Cnbp | 12321 | -0.203 | -0.1981 | Yes |
| 60 | Prdx5 | 12367 | -0.205 | -0.1910 | Yes |
| 61 | Sema3c | 12522 | -0.215 | -0.1900 | Yes |
| 62 | Cdk7 | 12599 | -0.219 | -0.1841 | Yes |
| 63 | Abcd3 | 12720 | -0.227 | -0.1805 | Yes |
| 64 | Mlycd | 12873 | -0.235 | -0.1785 | Yes |
| 65 | Abcd2 | 12964 | -0.241 | -0.1724 | Yes |
| 66 | Acsl1 | 13168 | -0.254 | -0.1725 | Yes |
| 67 | Hras | 13319 | -0.262 | -0.1690 | Yes |
| 68 | Pex14 | 13382 | -0.267 | -0.1600 | Yes |
| 69 | Abcd1 | 13551 | -0.278 | -0.1569 | Yes |
| 70 | Tspo | 13792 | -0.295 | -0.1572 | Yes |
| 71 | Hsd17b11 | 14147 | -0.317 | -0.1634 | Yes |
| 72 | Acaa1a | 14391 | -0.338 | -0.1619 | Yes |
| 73 | Nudt19 | 14451 | -0.344 | -0.1491 | Yes |
| 74 | Slc35b2 | 14539 | -0.351 | -0.1375 | Yes |
| 75 | Sult2b1 | 14751 | -0.369 | -0.1326 | Yes |
| 76 | Cacna1b | 14816 | -0.375 | -0.1186 | Yes |
| 77 | Bcl10 | 15035 | -0.397 | -0.1127 | Yes |
| 78 | Slc25a17 | 15221 | -0.415 | -0.1041 | Yes |
| 79 | Ehhadh | 15247 | -0.419 | -0.0856 | Yes |
| 80 | Ephx2 | 15356 | -0.433 | -0.0714 | Yes |
| 81 | Pex13 | 15838 | -0.504 | -0.0763 | Yes |
| 82 | Pex2 | 16096 | -0.563 | -0.0649 | Yes |
| 83 | Idh2 | 16257 | -0.608 | -0.0455 | Yes |
| 84 | Pex11b | 16470 | -0.718 | -0.0240 | Yes |
| 85 | Sod1 | 16556 | -0.791 | 0.0086 | Yes |