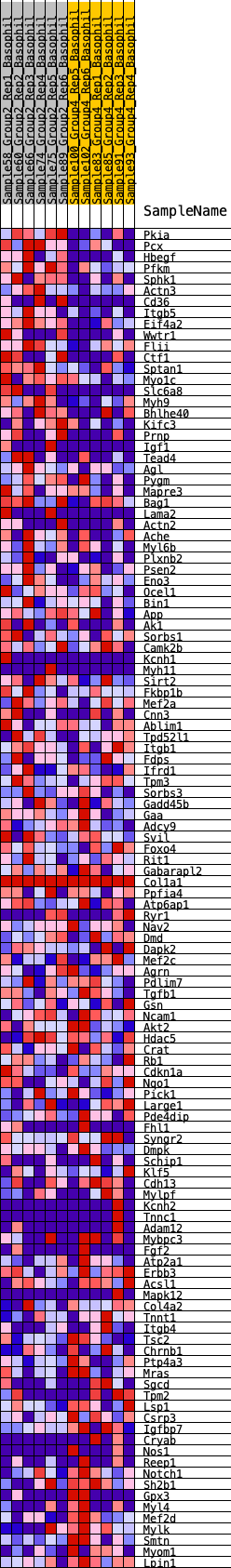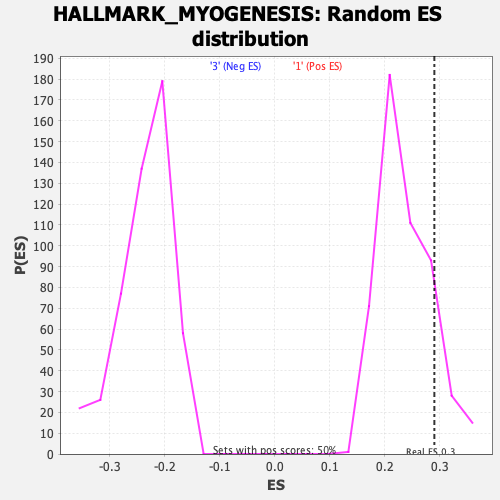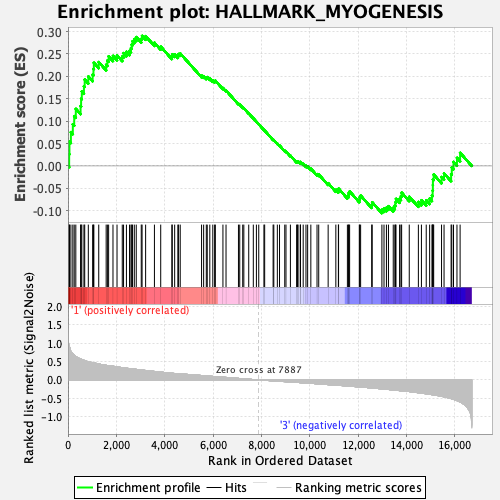
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.2903415 |
| Normalized Enrichment Score (NES) | 1.2328998 |
| Nominal p-value | 0.15369262 |
| FDR q-value | 0.78157383 |
| FWER p-Value | 0.923 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pkia | 48 | 0.927 | 0.0264 | Yes |
| 2 | Pcx | 60 | 0.907 | 0.0544 | Yes |
| 3 | Hbegf | 120 | 0.779 | 0.0755 | Yes |
| 4 | Pfkm | 200 | 0.708 | 0.0931 | Yes |
| 5 | Sphk1 | 257 | 0.678 | 0.1112 | Yes |
| 6 | Actn3 | 323 | 0.637 | 0.1274 | Yes |
| 7 | Cd36 | 520 | 0.570 | 0.1336 | Yes |
| 8 | Itgb5 | 542 | 0.565 | 0.1502 | Yes |
| 9 | Eif4a2 | 571 | 0.560 | 0.1662 | Yes |
| 10 | Wwtr1 | 659 | 0.537 | 0.1779 | Yes |
| 11 | Flii | 691 | 0.529 | 0.1928 | Yes |
| 12 | Ctf1 | 840 | 0.499 | 0.1996 | Yes |
| 13 | Sptan1 | 1019 | 0.471 | 0.2038 | Yes |
| 14 | Myo1c | 1053 | 0.466 | 0.2165 | Yes |
| 15 | Slc6a8 | 1069 | 0.463 | 0.2303 | Yes |
| 16 | Myh9 | 1271 | 0.435 | 0.2319 | Yes |
| 17 | Bhlhe40 | 1578 | 0.399 | 0.2260 | Yes |
| 18 | Kifc3 | 1629 | 0.393 | 0.2354 | Yes |
| 19 | Prnp | 1680 | 0.388 | 0.2447 | Yes |
| 20 | Igf1 | 1860 | 0.373 | 0.2457 | Yes |
| 21 | Tead4 | 2027 | 0.357 | 0.2470 | Yes |
| 22 | Agl | 2244 | 0.335 | 0.2445 | Yes |
| 23 | Pygm | 2297 | 0.331 | 0.2518 | Yes |
| 24 | Mapre3 | 2416 | 0.321 | 0.2549 | Yes |
| 25 | Bag1 | 2545 | 0.309 | 0.2569 | Yes |
| 26 | Lama2 | 2604 | 0.304 | 0.2631 | Yes |
| 27 | Actn2 | 2635 | 0.303 | 0.2708 | Yes |
| 28 | Ache | 2669 | 0.300 | 0.2783 | Yes |
| 29 | Myl6b | 2742 | 0.295 | 0.2833 | Yes |
| 30 | Plxnb2 | 2822 | 0.288 | 0.2877 | Yes |
| 31 | Psen2 | 3029 | 0.272 | 0.2838 | Yes |
| 32 | Eno3 | 3063 | 0.269 | 0.2903 | Yes |
| 33 | Ocel1 | 3210 | 0.258 | 0.2897 | No |
| 34 | Bin1 | 3574 | 0.232 | 0.2751 | No |
| 35 | App | 3834 | 0.214 | 0.2663 | No |
| 36 | Ak1 | 4296 | 0.183 | 0.2443 | No |
| 37 | Sorbs1 | 4310 | 0.183 | 0.2493 | No |
| 38 | Camk2b | 4411 | 0.176 | 0.2488 | No |
| 39 | Kcnh1 | 4543 | 0.169 | 0.2463 | No |
| 40 | Myh11 | 4573 | 0.168 | 0.2498 | No |
| 41 | Sirt2 | 4641 | 0.167 | 0.2511 | No |
| 42 | Fkbp1b | 5523 | 0.121 | 0.2018 | No |
| 43 | Mef2a | 5609 | 0.115 | 0.2003 | No |
| 44 | Cnn3 | 5719 | 0.110 | 0.1972 | No |
| 45 | Ablim1 | 5758 | 0.108 | 0.1983 | No |
| 46 | Tpd52l1 | 5864 | 0.102 | 0.1952 | No |
| 47 | Itgb1 | 5988 | 0.096 | 0.1908 | No |
| 48 | Fdps | 6063 | 0.093 | 0.1893 | No |
| 49 | Ifrd1 | 6093 | 0.091 | 0.1904 | No |
| 50 | Tpm3 | 6407 | 0.075 | 0.1739 | No |
| 51 | Sorbs3 | 6535 | 0.069 | 0.1684 | No |
| 52 | Gadd45b | 7049 | 0.043 | 0.1388 | No |
| 53 | Gaa | 7102 | 0.040 | 0.1369 | No |
| 54 | Adcy9 | 7225 | 0.033 | 0.1306 | No |
| 55 | Svil | 7271 | 0.031 | 0.1289 | No |
| 56 | Foxo4 | 7474 | 0.020 | 0.1173 | No |
| 57 | Rit1 | 7663 | 0.011 | 0.1063 | No |
| 58 | Gabarapl2 | 7791 | 0.005 | 0.0988 | No |
| 59 | Col1a1 | 7888 | 0.000 | 0.0930 | No |
| 60 | Ppfia4 | 8102 | -0.007 | 0.0804 | No |
| 61 | Atp6ap1 | 8133 | -0.008 | 0.0788 | No |
| 62 | Ryr1 | 8482 | -0.022 | 0.0585 | No |
| 63 | Nav2 | 8511 | -0.024 | 0.0576 | No |
| 64 | Dmd | 8657 | -0.030 | 0.0498 | No |
| 65 | Dapk2 | 8747 | -0.035 | 0.0455 | No |
| 66 | Mef2c | 8963 | -0.043 | 0.0339 | No |
| 67 | Agrn | 9014 | -0.045 | 0.0323 | No |
| 68 | Pdlim7 | 9197 | -0.053 | 0.0230 | No |
| 69 | Tgfb1 | 9454 | -0.064 | 0.0096 | No |
| 70 | Gsn | 9489 | -0.066 | 0.0096 | No |
| 71 | Ncam1 | 9514 | -0.067 | 0.0103 | No |
| 72 | Akt2 | 9604 | -0.071 | 0.0072 | No |
| 73 | Hdac5 | 9614 | -0.072 | 0.0089 | No |
| 74 | Crat | 9726 | -0.077 | 0.0046 | No |
| 75 | Rb1 | 9841 | -0.082 | 0.0003 | No |
| 76 | Cdkn1a | 9910 | -0.084 | -0.0011 | No |
| 77 | Nqo1 | 10042 | -0.091 | -0.0061 | No |
| 78 | Pick1 | 10298 | -0.102 | -0.0183 | No |
| 79 | Large1 | 10368 | -0.106 | -0.0191 | No |
| 80 | Pde4dip | 10758 | -0.125 | -0.0386 | No |
| 81 | Fhl1 | 11076 | -0.142 | -0.0532 | No |
| 82 | Syngr2 | 11181 | -0.146 | -0.0549 | No |
| 83 | Dmpk | 11189 | -0.146 | -0.0507 | No |
| 84 | Schip1 | 11554 | -0.163 | -0.0675 | No |
| 85 | Klf5 | 11603 | -0.167 | -0.0651 | No |
| 86 | Cdh13 | 11611 | -0.167 | -0.0602 | No |
| 87 | Mylpf | 11646 | -0.169 | -0.0569 | No |
| 88 | Kcnh2 | 12046 | -0.189 | -0.0750 | No |
| 89 | Tnnc1 | 12061 | -0.189 | -0.0699 | No |
| 90 | Adam12 | 12101 | -0.191 | -0.0662 | No |
| 91 | Mybpc3 | 12559 | -0.217 | -0.0869 | No |
| 92 | Fgf2 | 12579 | -0.218 | -0.0812 | No |
| 93 | Atp2a1 | 12981 | -0.242 | -0.0977 | No |
| 94 | Erbb3 | 13068 | -0.247 | -0.0951 | No |
| 95 | Acsl1 | 13168 | -0.254 | -0.0930 | No |
| 96 | Mapk12 | 13252 | -0.258 | -0.0899 | No |
| 97 | Col4a2 | 13450 | -0.271 | -0.0932 | No |
| 98 | Tnnt1 | 13515 | -0.276 | -0.0883 | No |
| 99 | Itgb4 | 13542 | -0.278 | -0.0811 | No |
| 100 | Tsc2 | 13563 | -0.279 | -0.0735 | No |
| 101 | Chrnb1 | 13712 | -0.289 | -0.0733 | No |
| 102 | Ptp4a3 | 13768 | -0.293 | -0.0673 | No |
| 103 | Mras | 13797 | -0.295 | -0.0597 | No |
| 104 | Sgcd | 14112 | -0.314 | -0.0687 | No |
| 105 | Tpm2 | 14489 | -0.347 | -0.0804 | No |
| 106 | Lsp1 | 14615 | -0.357 | -0.0766 | No |
| 107 | Csrp3 | 14814 | -0.375 | -0.0767 | No |
| 108 | Igfbp7 | 14955 | -0.388 | -0.0729 | No |
| 109 | Cryab | 15051 | -0.399 | -0.0660 | No |
| 110 | Nos1 | 15080 | -0.400 | -0.0550 | No |
| 111 | Reep1 | 15088 | -0.401 | -0.0428 | No |
| 112 | Notch1 | 15091 | -0.401 | -0.0302 | No |
| 113 | Sh2b1 | 15122 | -0.405 | -0.0193 | No |
| 114 | Gpx3 | 15447 | -0.444 | -0.0248 | No |
| 115 | Myl4 | 15551 | -0.456 | -0.0166 | No |
| 116 | Mef2d | 15843 | -0.506 | -0.0181 | No |
| 117 | Mylk | 15871 | -0.513 | -0.0035 | No |
| 118 | Smtn | 15942 | -0.527 | 0.0089 | No |
| 119 | Myom1 | 16085 | -0.561 | 0.0181 | No |
| 120 | Lpin1 | 16215 | -0.598 | 0.0292 | No |