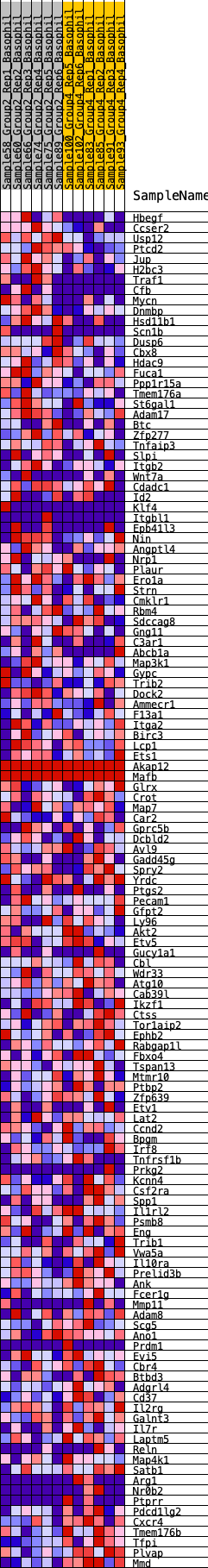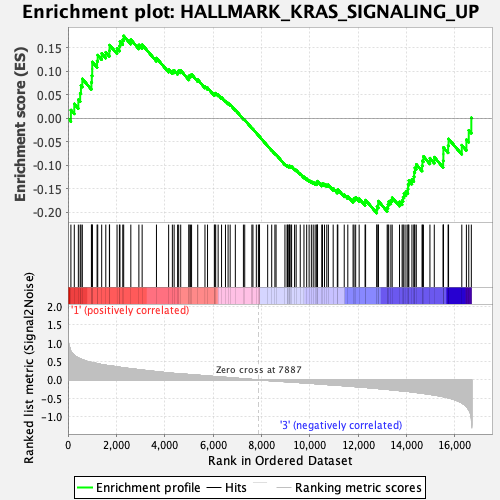
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
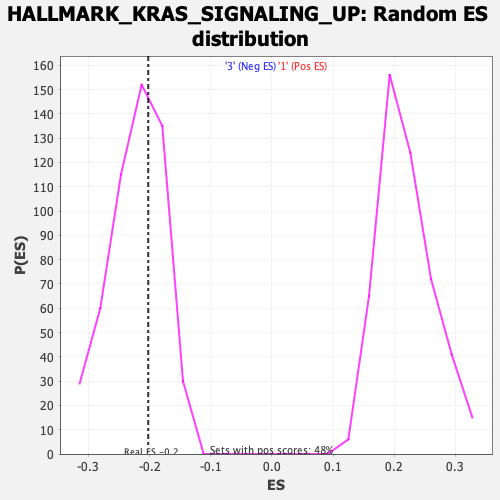
| Enrichment Score (ES) | -0.20231372 |
| Normalized Enrichment Score (NES) | -0.91801065 |
| Nominal p-value | 0.59117085 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hbegf | 120 | 0.779 | 0.0176 | No |
| 2 | Ccser2 | 260 | 0.674 | 0.0307 | No |
| 3 | Usp12 | 428 | 0.599 | 0.0397 | No |
| 4 | Ptcd2 | 503 | 0.574 | 0.0536 | No |
| 5 | Jup | 536 | 0.567 | 0.0697 | No |
| 6 | H2bc3 | 592 | 0.553 | 0.0840 | No |
| 7 | Traf1 | 968 | 0.477 | 0.0766 | No |
| 8 | Cfb | 985 | 0.475 | 0.0908 | No |
| 9 | Mycn | 1001 | 0.473 | 0.1050 | No |
| 10 | Dnmbp | 1002 | 0.473 | 0.1201 | No |
| 11 | Hsd11b1 | 1203 | 0.446 | 0.1222 | No |
| 12 | Scn1b | 1229 | 0.440 | 0.1348 | No |
| 13 | Dusp6 | 1393 | 0.418 | 0.1383 | No |
| 14 | Cbx8 | 1563 | 0.400 | 0.1408 | No |
| 15 | Hdac9 | 1712 | 0.385 | 0.1442 | No |
| 16 | Fuca1 | 1719 | 0.385 | 0.1561 | No |
| 17 | Ppp1r15a | 2028 | 0.357 | 0.1488 | No |
| 18 | Tmem176a | 2126 | 0.346 | 0.1540 | No |
| 19 | St6gal1 | 2148 | 0.344 | 0.1637 | No |
| 20 | Adam17 | 2265 | 0.333 | 0.1674 | No |
| 21 | Btc | 2303 | 0.330 | 0.1757 | No |
| 22 | Zfp277 | 2598 | 0.305 | 0.1676 | No |
| 23 | Tnfaip3 | 2931 | 0.279 | 0.1565 | No |
| 24 | Slpi | 3066 | 0.269 | 0.1570 | No |
| 25 | Itgb2 | 3658 | 0.225 | 0.1285 | No |
| 26 | Wnt7a | 4162 | 0.192 | 0.1042 | No |
| 27 | Cdadc1 | 4316 | 0.182 | 0.1008 | No |
| 28 | Id2 | 4389 | 0.178 | 0.1021 | No |
| 29 | Klf4 | 4533 | 0.169 | 0.0989 | No |
| 30 | Itgbl1 | 4577 | 0.168 | 0.1017 | No |
| 31 | Epb41l3 | 4661 | 0.166 | 0.1019 | No |
| 32 | Nin | 5001 | 0.147 | 0.0862 | No |
| 33 | Angptl4 | 5005 | 0.147 | 0.0907 | No |
| 34 | Nrp1 | 5063 | 0.144 | 0.0918 | No |
| 35 | Plaur | 5110 | 0.141 | 0.0936 | No |
| 36 | Ero1a | 5365 | 0.128 | 0.0823 | No |
| 37 | Strn | 5664 | 0.112 | 0.0679 | No |
| 38 | Cmklr1 | 5770 | 0.107 | 0.0650 | No |
| 39 | Rbm4 | 6052 | 0.093 | 0.0510 | No |
| 40 | Sdccag8 | 6074 | 0.092 | 0.0527 | No |
| 41 | Gng11 | 6107 | 0.091 | 0.0536 | No |
| 42 | C3ar1 | 6209 | 0.085 | 0.0502 | No |
| 43 | Abcb1a | 6350 | 0.078 | 0.0443 | No |
| 44 | Map3k1 | 6512 | 0.070 | 0.0368 | No |
| 45 | Gypc | 6620 | 0.065 | 0.0324 | No |
| 46 | Trib2 | 6706 | 0.060 | 0.0292 | No |
| 47 | Dock2 | 6920 | 0.048 | 0.0179 | No |
| 48 | Ammecr1 | 7254 | 0.031 | -0.0012 | No |
| 49 | F13a1 | 7308 | 0.028 | -0.0035 | No |
| 50 | Itga2 | 7605 | 0.014 | -0.0209 | No |
| 51 | Birc3 | 7648 | 0.012 | -0.0231 | No |
| 52 | Lcp1 | 7788 | 0.005 | -0.0313 | No |
| 53 | Ets1 | 7873 | 0.001 | -0.0364 | No |
| 54 | Akap12 | 7904 | 0.000 | -0.0382 | No |
| 55 | Mafb | 7920 | 0.000 | -0.0391 | No |
| 56 | Glrx | 8257 | -0.013 | -0.0590 | No |
| 57 | Crot | 8424 | -0.020 | -0.0684 | No |
| 58 | Map7 | 8556 | -0.025 | -0.0755 | No |
| 59 | Car2 | 8604 | -0.028 | -0.0774 | No |
| 60 | Gprc5b | 8971 | -0.043 | -0.0981 | No |
| 61 | Dcbld2 | 9060 | -0.047 | -0.1019 | No |
| 62 | Avl9 | 9076 | -0.048 | -0.1013 | No |
| 63 | Gadd45g | 9131 | -0.049 | -0.1030 | No |
| 64 | Spry2 | 9152 | -0.051 | -0.1026 | No |
| 65 | Yrdc | 9162 | -0.051 | -0.1015 | No |
| 66 | Ptgs2 | 9217 | -0.053 | -0.1031 | No |
| 67 | Pecam1 | 9247 | -0.054 | -0.1031 | No |
| 68 | Gfpt2 | 9370 | -0.060 | -0.1085 | No |
| 69 | Ly96 | 9435 | -0.063 | -0.1104 | No |
| 70 | Akt2 | 9604 | -0.071 | -0.1182 | No |
| 71 | Etv5 | 9758 | -0.078 | -0.1250 | No |
| 72 | Gucy1a1 | 9870 | -0.083 | -0.1290 | No |
| 73 | Cbl | 9971 | -0.088 | -0.1323 | No |
| 74 | Wdr33 | 10063 | -0.091 | -0.1348 | No |
| 75 | Atg10 | 10140 | -0.095 | -0.1364 | No |
| 76 | Cab39l | 10220 | -0.098 | -0.1380 | No |
| 77 | Ikzf1 | 10285 | -0.101 | -0.1387 | No |
| 78 | Ctss | 10305 | -0.102 | -0.1366 | No |
| 79 | Tor1aip2 | 10319 | -0.103 | -0.1340 | No |
| 80 | Ephb2 | 10495 | -0.113 | -0.1410 | No |
| 81 | Rabgap1l | 10523 | -0.114 | -0.1390 | No |
| 82 | Fbxo4 | 10608 | -0.118 | -0.1403 | No |
| 83 | Tspan13 | 10701 | -0.122 | -0.1420 | No |
| 84 | Mtmr10 | 10767 | -0.126 | -0.1419 | No |
| 85 | Ptbp2 | 10966 | -0.136 | -0.1495 | No |
| 86 | Zfp639 | 11137 | -0.144 | -0.1551 | No |
| 87 | Etv1 | 11158 | -0.145 | -0.1517 | No |
| 88 | Lat2 | 11423 | -0.156 | -0.1627 | No |
| 89 | Ccnd2 | 11571 | -0.165 | -0.1663 | No |
| 90 | Bpgm | 11789 | -0.175 | -0.1738 | No |
| 91 | Irf8 | 11829 | -0.177 | -0.1705 | No |
| 92 | Tnfrsf1b | 11896 | -0.180 | -0.1688 | No |
| 93 | Prkg2 | 12040 | -0.189 | -0.1714 | No |
| 94 | Kcnn4 | 12285 | -0.201 | -0.1797 | No |
| 95 | Csf2ra | 12307 | -0.202 | -0.1745 | No |
| 96 | Spp1 | 12769 | -0.230 | -0.1950 | Yes |
| 97 | Il1rl2 | 12778 | -0.230 | -0.1882 | Yes |
| 98 | Psmb8 | 12832 | -0.233 | -0.1839 | Yes |
| 99 | Eng | 12834 | -0.233 | -0.1766 | Yes |
| 100 | Trib1 | 13199 | -0.255 | -0.1904 | Yes |
| 101 | Vwa5a | 13232 | -0.257 | -0.1841 | Yes |
| 102 | Il10ra | 13263 | -0.259 | -0.1777 | Yes |
| 103 | Prelid3b | 13352 | -0.264 | -0.1746 | Yes |
| 104 | Ank | 13406 | -0.268 | -0.1692 | Yes |
| 105 | Fcer1g | 13709 | -0.289 | -0.1782 | Yes |
| 106 | Mmp11 | 13817 | -0.297 | -0.1752 | Yes |
| 107 | Adam8 | 13857 | -0.299 | -0.1680 | Yes |
| 108 | Scg5 | 13894 | -0.301 | -0.1606 | Yes |
| 109 | Ano1 | 13976 | -0.307 | -0.1557 | Yes |
| 110 | Prdm1 | 14057 | -0.311 | -0.1506 | Yes |
| 111 | Evi5 | 14065 | -0.311 | -0.1411 | Yes |
| 112 | Cbr4 | 14096 | -0.313 | -0.1329 | Yes |
| 113 | Btbd3 | 14223 | -0.324 | -0.1302 | Yes |
| 114 | Adgrl4 | 14303 | -0.330 | -0.1245 | Yes |
| 115 | Cd37 | 14317 | -0.332 | -0.1147 | Yes |
| 116 | Il2rg | 14345 | -0.334 | -0.1057 | Yes |
| 117 | Galnt3 | 14403 | -0.339 | -0.0983 | Yes |
| 118 | Il7r | 14643 | -0.360 | -0.1012 | Yes |
| 119 | Laptm5 | 14657 | -0.361 | -0.0905 | Yes |
| 120 | Reln | 14698 | -0.364 | -0.0813 | Yes |
| 121 | Map4k1 | 14967 | -0.390 | -0.0851 | Yes |
| 122 | Satb1 | 15147 | -0.407 | -0.0829 | Yes |
| 123 | Arg1 | 15514 | -0.452 | -0.0905 | Yes |
| 124 | Nr0b2 | 15523 | -0.452 | -0.0766 | Yes |
| 125 | Ptprr | 15524 | -0.452 | -0.0622 | Yes |
| 126 | Pdcd1lg2 | 15719 | -0.483 | -0.0585 | Yes |
| 127 | Cxcr4 | 15737 | -0.486 | -0.0440 | Yes |
| 128 | Tmem176b | 16285 | -0.621 | -0.0572 | Yes |
| 129 | Tfpi | 16475 | -0.723 | -0.0456 | Yes |
| 130 | Plvap | 16573 | -0.808 | -0.0257 | Yes |
| 131 | Mmd | 16679 | -1.042 | 0.0012 | Yes |