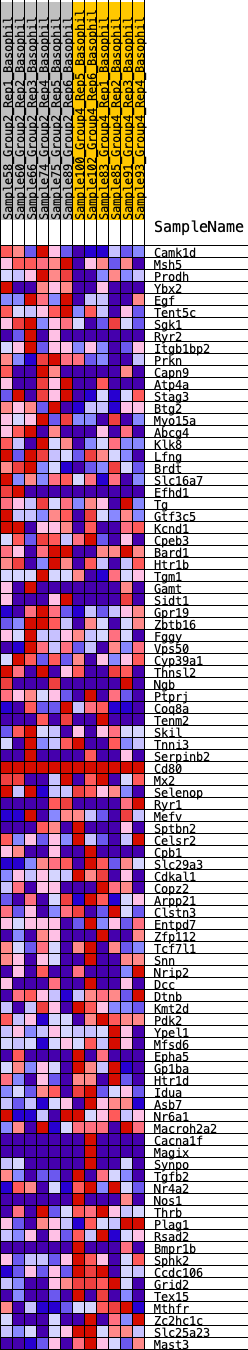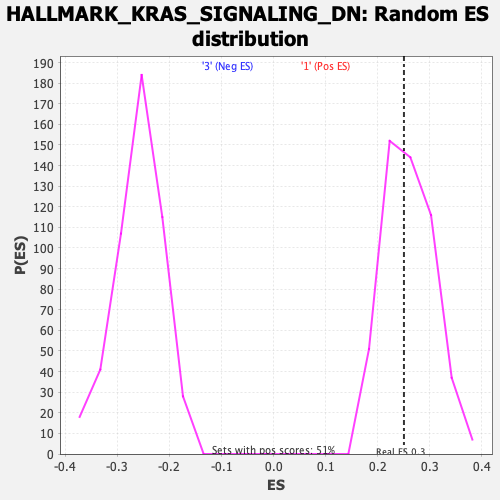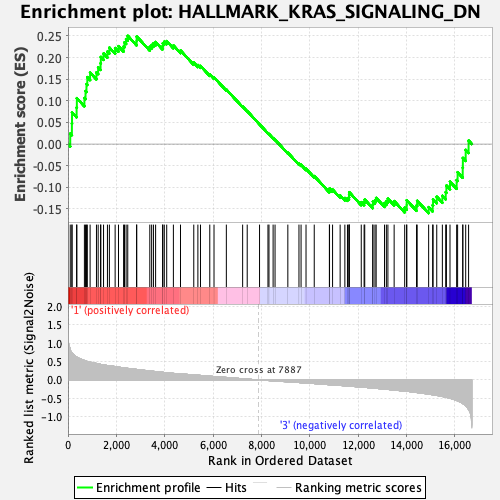
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.25028494 |
| Normalized Enrichment Score (NES) | 0.9682633 |
| Nominal p-value | 0.54043394 |
| FDR q-value | 0.6479121 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Camk1d | 93 | 0.832 | 0.0244 | Yes |
| 2 | Msh5 | 161 | 0.742 | 0.0471 | Yes |
| 3 | Prodh | 167 | 0.733 | 0.0732 | Yes |
| 4 | Ybx2 | 356 | 0.624 | 0.0844 | Yes |
| 5 | Egf | 366 | 0.620 | 0.1062 | Yes |
| 6 | Tent5c | 678 | 0.532 | 0.1066 | Yes |
| 7 | Sgk1 | 725 | 0.521 | 0.1226 | Yes |
| 8 | Ryr2 | 764 | 0.509 | 0.1387 | Yes |
| 9 | Itgb1bp2 | 799 | 0.504 | 0.1548 | Yes |
| 10 | Prkn | 914 | 0.486 | 0.1654 | Yes |
| 11 | Capn9 | 1177 | 0.450 | 0.1659 | Yes |
| 12 | Atp4a | 1247 | 0.438 | 0.1775 | Yes |
| 13 | Stag3 | 1347 | 0.424 | 0.1868 | Yes |
| 14 | Btg2 | 1357 | 0.423 | 0.2015 | Yes |
| 15 | Myo15a | 1470 | 0.410 | 0.2095 | Yes |
| 16 | Abcg4 | 1631 | 0.393 | 0.2140 | Yes |
| 17 | Klk8 | 1711 | 0.385 | 0.2231 | Yes |
| 18 | Lfng | 1949 | 0.367 | 0.2221 | Yes |
| 19 | Brdt | 2088 | 0.351 | 0.2264 | Yes |
| 20 | Slc16a7 | 2299 | 0.331 | 0.2257 | Yes |
| 21 | Efhd1 | 2336 | 0.327 | 0.2353 | Yes |
| 22 | Tg | 2403 | 0.322 | 0.2429 | Yes |
| 23 | Gtf3c5 | 2471 | 0.316 | 0.2503 | Yes |
| 24 | Kcnd1 | 2838 | 0.286 | 0.2386 | No |
| 25 | Cpeb3 | 2840 | 0.286 | 0.2488 | No |
| 26 | Bard1 | 3383 | 0.246 | 0.2250 | No |
| 27 | Htr1b | 3459 | 0.240 | 0.2292 | No |
| 28 | Tgm1 | 3533 | 0.235 | 0.2332 | No |
| 29 | Gamt | 3623 | 0.228 | 0.2361 | No |
| 30 | Sidt1 | 3911 | 0.209 | 0.2263 | No |
| 31 | Gpr19 | 3922 | 0.208 | 0.2332 | No |
| 32 | Zbtb16 | 3982 | 0.203 | 0.2370 | No |
| 33 | Fggy | 4082 | 0.197 | 0.2381 | No |
| 34 | Vps50 | 4358 | 0.179 | 0.2280 | No |
| 35 | Cyp39a1 | 4652 | 0.167 | 0.2164 | No |
| 36 | Thnsl2 | 5201 | 0.136 | 0.1883 | No |
| 37 | Ngb | 5371 | 0.128 | 0.1827 | No |
| 38 | Ptprj | 5479 | 0.123 | 0.1807 | No |
| 39 | Coq8a | 5863 | 0.102 | 0.1613 | No |
| 40 | Tenm2 | 6038 | 0.094 | 0.1542 | No |
| 41 | Skil | 6548 | 0.068 | 0.1261 | No |
| 42 | Tnni3 | 7222 | 0.033 | 0.0867 | No |
| 43 | Serpinb2 | 7411 | 0.023 | 0.0762 | No |
| 44 | Cd80 | 7921 | 0.000 | 0.0456 | No |
| 45 | Mx2 | 8269 | -0.013 | 0.0252 | No |
| 46 | Selenop | 8311 | -0.015 | 0.0232 | No |
| 47 | Ryr1 | 8482 | -0.022 | 0.0138 | No |
| 48 | Mefv | 8562 | -0.026 | 0.0100 | No |
| 49 | Sptbn2 | 9091 | -0.048 | -0.0201 | No |
| 50 | Celsr2 | 9551 | -0.068 | -0.0453 | No |
| 51 | Cpb1 | 9638 | -0.073 | -0.0478 | No |
| 52 | Slc29a3 | 9843 | -0.082 | -0.0572 | No |
| 53 | Cdkal1 | 10183 | -0.097 | -0.0741 | No |
| 54 | Copz2 | 10809 | -0.129 | -0.1071 | No |
| 55 | Arpp21 | 10815 | -0.129 | -0.1028 | No |
| 56 | Clstn3 | 10941 | -0.135 | -0.1054 | No |
| 57 | Entpd7 | 11254 | -0.149 | -0.1188 | No |
| 58 | Zfp112 | 11449 | -0.157 | -0.1248 | No |
| 59 | Tcf7l1 | 11557 | -0.164 | -0.1254 | No |
| 60 | Snn | 11618 | -0.167 | -0.1230 | No |
| 61 | Nrip2 | 11626 | -0.168 | -0.1174 | No |
| 62 | Dcc | 11630 | -0.168 | -0.1115 | No |
| 63 | Dtnb | 12127 | -0.192 | -0.1344 | No |
| 64 | Kmt2d | 12245 | -0.199 | -0.1343 | No |
| 65 | Pdk2 | 12271 | -0.200 | -0.1286 | No |
| 66 | Ypel1 | 12602 | -0.219 | -0.1406 | No |
| 67 | Mfsd6 | 12610 | -0.220 | -0.1331 | No |
| 68 | Epha5 | 12694 | -0.226 | -0.1299 | No |
| 69 | Gp1ba | 12743 | -0.228 | -0.1246 | No |
| 70 | Htr1d | 13089 | -0.249 | -0.1364 | No |
| 71 | Idua | 13165 | -0.254 | -0.1318 | No |
| 72 | Asb7 | 13229 | -0.257 | -0.1263 | No |
| 73 | Nr6a1 | 13487 | -0.274 | -0.1319 | No |
| 74 | Macroh2a2 | 13923 | -0.303 | -0.1472 | No |
| 75 | Cacna1f | 14006 | -0.308 | -0.1410 | No |
| 76 | Magix | 14007 | -0.308 | -0.1299 | No |
| 77 | Synpo | 14413 | -0.340 | -0.1421 | No |
| 78 | Tgfb2 | 14439 | -0.342 | -0.1312 | No |
| 79 | Nr4a2 | 14914 | -0.384 | -0.1459 | No |
| 80 | Nos1 | 15080 | -0.400 | -0.1415 | No |
| 81 | Thrb | 15097 | -0.402 | -0.1279 | No |
| 82 | Plag1 | 15249 | -0.419 | -0.1219 | No |
| 83 | Rsad2 | 15481 | -0.449 | -0.1197 | No |
| 84 | Bmpr1b | 15620 | -0.466 | -0.1112 | No |
| 85 | Sphk2 | 15649 | -0.470 | -0.0959 | No |
| 86 | Ccdc106 | 15797 | -0.496 | -0.0869 | No |
| 87 | Grid2 | 16072 | -0.559 | -0.0833 | No |
| 88 | Tex15 | 16112 | -0.568 | -0.0652 | No |
| 89 | Mthfr | 16323 | -0.637 | -0.0549 | No |
| 90 | Zc2hc1c | 16329 | -0.641 | -0.0321 | No |
| 91 | Slc25a23 | 16446 | -0.708 | -0.0136 | No |
| 92 | Mast3 | 16566 | -0.797 | 0.0080 | No |