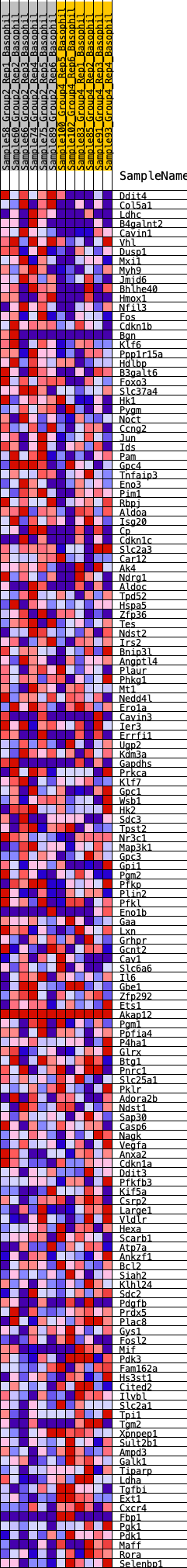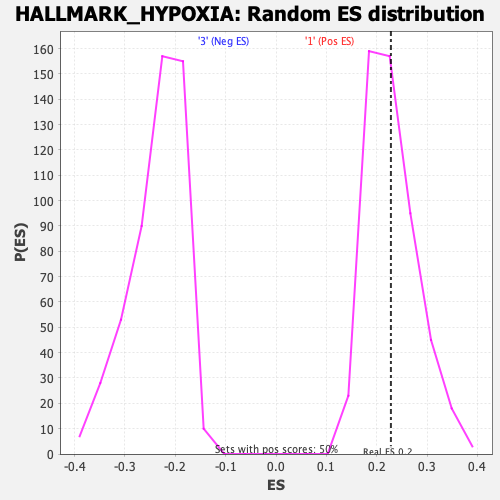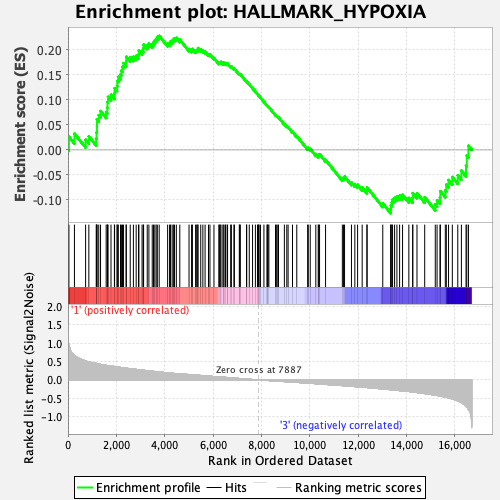
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.22828084 |
| Normalized Enrichment Score (NES) | 0.9896529 |
| Nominal p-value | 0.468 |
| FDR q-value | 0.62635267 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ddit4 | 37 | 0.962 | 0.0265 | Yes |
| 2 | Col5a1 | 266 | 0.667 | 0.0327 | Yes |
| 3 | Ldhc | 728 | 0.521 | 0.0205 | Yes |
| 4 | B4galnt2 | 862 | 0.495 | 0.0272 | Yes |
| 5 | Cavin1 | 1167 | 0.451 | 0.0223 | Yes |
| 6 | Vhl | 1181 | 0.450 | 0.0350 | Yes |
| 7 | Dusp1 | 1195 | 0.447 | 0.0476 | Yes |
| 8 | Mxi1 | 1197 | 0.447 | 0.0609 | Yes |
| 9 | Myh9 | 1271 | 0.435 | 0.0695 | Yes |
| 10 | Jmjd6 | 1345 | 0.424 | 0.0778 | Yes |
| 11 | Bhlhe40 | 1578 | 0.399 | 0.0757 | Yes |
| 12 | Hmox1 | 1627 | 0.393 | 0.0845 | Yes |
| 13 | Nfil3 | 1632 | 0.392 | 0.0960 | Yes |
| 14 | Fos | 1657 | 0.391 | 0.1063 | Yes |
| 15 | Cdkn1b | 1778 | 0.379 | 0.1104 | Yes |
| 16 | Bgn | 1919 | 0.368 | 0.1129 | Yes |
| 17 | Klf6 | 1927 | 0.368 | 0.1235 | Yes |
| 18 | Ppp1r15a | 2028 | 0.357 | 0.1282 | Yes |
| 19 | Hdlbp | 2039 | 0.355 | 0.1382 | Yes |
| 20 | B3galt6 | 2081 | 0.352 | 0.1462 | Yes |
| 21 | Foxo3 | 2171 | 0.342 | 0.1511 | Yes |
| 22 | Slc37a4 | 2209 | 0.338 | 0.1589 | Yes |
| 23 | Hk1 | 2254 | 0.334 | 0.1663 | Yes |
| 24 | Pygm | 2297 | 0.331 | 0.1736 | Yes |
| 25 | Noct | 2406 | 0.322 | 0.1768 | Yes |
| 26 | Ccng2 | 2408 | 0.322 | 0.1863 | Yes |
| 27 | Jun | 2573 | 0.307 | 0.1856 | Yes |
| 28 | Ids | 2704 | 0.298 | 0.1867 | Yes |
| 29 | Pam | 2819 | 0.289 | 0.1884 | Yes |
| 30 | Gpc4 | 2917 | 0.280 | 0.1909 | Yes |
| 31 | Tnfaip3 | 2931 | 0.279 | 0.1985 | Yes |
| 32 | Eno3 | 3063 | 0.269 | 0.1986 | Yes |
| 33 | Pim1 | 3113 | 0.265 | 0.2036 | Yes |
| 34 | Rbpj | 3126 | 0.264 | 0.2107 | Yes |
| 35 | Aldoa | 3277 | 0.254 | 0.2093 | Yes |
| 36 | Isg20 | 3345 | 0.249 | 0.2127 | Yes |
| 37 | Cp | 3483 | 0.239 | 0.2116 | Yes |
| 38 | Cdkn1c | 3536 | 0.235 | 0.2154 | Yes |
| 39 | Slc2a3 | 3590 | 0.231 | 0.2191 | Yes |
| 40 | Car12 | 3646 | 0.226 | 0.2226 | Yes |
| 41 | Ak4 | 3694 | 0.222 | 0.2264 | Yes |
| 42 | Ndrg1 | 3771 | 0.217 | 0.2283 | Yes |
| 43 | Aldoc | 4120 | 0.195 | 0.2131 | No |
| 44 | Tpd52 | 4200 | 0.189 | 0.2140 | No |
| 45 | Hspa5 | 4240 | 0.187 | 0.2172 | No |
| 46 | Zfp36 | 4306 | 0.183 | 0.2187 | No |
| 47 | Tes | 4357 | 0.179 | 0.2211 | No |
| 48 | Ndst2 | 4405 | 0.176 | 0.2235 | No |
| 49 | Irs2 | 4480 | 0.172 | 0.2242 | No |
| 50 | Bnip3l | 4623 | 0.168 | 0.2206 | No |
| 51 | Angptl4 | 5005 | 0.147 | 0.2020 | No |
| 52 | Plaur | 5110 | 0.141 | 0.2000 | No |
| 53 | Phkg1 | 5145 | 0.140 | 0.2021 | No |
| 54 | Mt1 | 5276 | 0.132 | 0.1982 | No |
| 55 | Nedd4l | 5322 | 0.130 | 0.1994 | No |
| 56 | Ero1a | 5365 | 0.128 | 0.2006 | No |
| 57 | Cavin3 | 5376 | 0.128 | 0.2039 | No |
| 58 | Ier3 | 5489 | 0.123 | 0.2008 | No |
| 59 | Errfi1 | 5577 | 0.117 | 0.1990 | No |
| 60 | Ugp2 | 5669 | 0.112 | 0.1969 | No |
| 61 | Kdm3a | 5810 | 0.105 | 0.1916 | No |
| 62 | Gapdhs | 5870 | 0.101 | 0.1910 | No |
| 63 | Prkca | 6036 | 0.094 | 0.1839 | No |
| 64 | Klf7 | 6238 | 0.084 | 0.1742 | No |
| 65 | Gpc1 | 6261 | 0.083 | 0.1754 | No |
| 66 | Wsb1 | 6296 | 0.080 | 0.1757 | No |
| 67 | Hk2 | 6327 | 0.078 | 0.1762 | No |
| 68 | Sdc3 | 6414 | 0.074 | 0.1733 | No |
| 69 | Tpst2 | 6415 | 0.074 | 0.1755 | No |
| 70 | Nr3c1 | 6490 | 0.071 | 0.1732 | No |
| 71 | Map3k1 | 6512 | 0.070 | 0.1740 | No |
| 72 | Gpc3 | 6579 | 0.067 | 0.1720 | No |
| 73 | Gpi1 | 6599 | 0.066 | 0.1728 | No |
| 74 | Pgm2 | 6728 | 0.059 | 0.1669 | No |
| 75 | Pfkp | 6756 | 0.057 | 0.1669 | No |
| 76 | Plin2 | 6862 | 0.051 | 0.1621 | No |
| 77 | Pfkl | 6881 | 0.050 | 0.1626 | No |
| 78 | Eno1b | 7084 | 0.041 | 0.1516 | No |
| 79 | Gaa | 7102 | 0.040 | 0.1517 | No |
| 80 | Lxn | 7125 | 0.039 | 0.1516 | No |
| 81 | Grhpr | 7386 | 0.024 | 0.1366 | No |
| 82 | Gcnt2 | 7387 | 0.024 | 0.1373 | No |
| 83 | Cav1 | 7496 | 0.019 | 0.1314 | No |
| 84 | Slc6a6 | 7632 | 0.013 | 0.1236 | No |
| 85 | Il6 | 7749 | 0.007 | 0.1168 | No |
| 86 | Gbe1 | 7835 | 0.003 | 0.1117 | No |
| 87 | Zfp292 | 7868 | 0.001 | 0.1098 | No |
| 88 | Ets1 | 7873 | 0.001 | 0.1096 | No |
| 89 | Akap12 | 7904 | 0.000 | 0.1078 | No |
| 90 | Pgm1 | 7959 | -0.001 | 0.1045 | No |
| 91 | Ppfia4 | 8102 | -0.007 | 0.0962 | No |
| 92 | P4ha1 | 8227 | -0.012 | 0.0890 | No |
| 93 | Glrx | 8257 | -0.013 | 0.0877 | No |
| 94 | Btg1 | 8268 | -0.013 | 0.0874 | No |
| 95 | Pnrc1 | 8307 | -0.015 | 0.0856 | No |
| 96 | Slc25a1 | 8581 | -0.027 | 0.0699 | No |
| 97 | Pklr | 8629 | -0.029 | 0.0679 | No |
| 98 | Adora2b | 8678 | -0.031 | 0.0659 | No |
| 99 | Ndst1 | 8705 | -0.032 | 0.0653 | No |
| 100 | Sap30 | 8948 | -0.042 | 0.0520 | No |
| 101 | Casp6 | 9048 | -0.046 | 0.0474 | No |
| 102 | Nagk | 9102 | -0.048 | 0.0456 | No |
| 103 | Vegfa | 9284 | -0.057 | 0.0364 | No |
| 104 | Anxa2 | 9461 | -0.065 | 0.0277 | No |
| 105 | Cdkn1a | 9910 | -0.084 | 0.0032 | No |
| 106 | Ddit3 | 9936 | -0.086 | 0.0042 | No |
| 107 | Pfkfb3 | 10017 | -0.089 | 0.0021 | No |
| 108 | Kif5a | 10248 | -0.099 | -0.0089 | No |
| 109 | Csrp2 | 10347 | -0.105 | -0.0116 | No |
| 110 | Large1 | 10368 | -0.106 | -0.0097 | No |
| 111 | Vldlr | 10403 | -0.108 | -0.0085 | No |
| 112 | Hexa | 10651 | -0.120 | -0.0198 | No |
| 113 | Scarb1 | 11345 | -0.152 | -0.0572 | No |
| 114 | Atp7a | 11395 | -0.154 | -0.0555 | No |
| 115 | Ankzf1 | 11436 | -0.157 | -0.0532 | No |
| 116 | Bcl2 | 11722 | -0.172 | -0.0653 | No |
| 117 | Siah2 | 11862 | -0.179 | -0.0684 | No |
| 118 | Klhl24 | 11976 | -0.185 | -0.0697 | No |
| 119 | Sdc2 | 12165 | -0.195 | -0.0752 | No |
| 120 | Pdgfb | 12363 | -0.205 | -0.0809 | No |
| 121 | Prdx5 | 12367 | -0.205 | -0.0750 | No |
| 122 | Plac8 | 13017 | -0.245 | -0.1069 | No |
| 123 | Gys1 | 13342 | -0.264 | -0.1186 | No |
| 124 | Fosl2 | 13355 | -0.264 | -0.1114 | No |
| 125 | Mif | 13388 | -0.267 | -0.1053 | No |
| 126 | Pdk3 | 13422 | -0.269 | -0.0993 | No |
| 127 | Fam162a | 13506 | -0.275 | -0.0960 | No |
| 128 | Hs3st1 | 13602 | -0.282 | -0.0933 | No |
| 129 | Cited2 | 13713 | -0.289 | -0.0913 | No |
| 130 | Ilvbl | 13835 | -0.297 | -0.0897 | No |
| 131 | Slc2a1 | 14095 | -0.313 | -0.0960 | No |
| 132 | Tpi1 | 14253 | -0.326 | -0.0958 | No |
| 133 | Tgm2 | 14259 | -0.326 | -0.0863 | No |
| 134 | Xpnpep1 | 14432 | -0.342 | -0.0865 | No |
| 135 | Sult2b1 | 14751 | -0.369 | -0.0946 | No |
| 136 | Ampd3 | 15188 | -0.412 | -0.1086 | No |
| 137 | Galk1 | 15262 | -0.421 | -0.1005 | No |
| 138 | Tiparp | 15384 | -0.436 | -0.0947 | No |
| 139 | Ldha | 15399 | -0.439 | -0.0824 | No |
| 140 | Tgfbi | 15603 | -0.464 | -0.0808 | No |
| 141 | Ext1 | 15645 | -0.469 | -0.0693 | No |
| 142 | Cxcr4 | 15737 | -0.486 | -0.0602 | No |
| 143 | Fbp1 | 15893 | -0.516 | -0.0542 | No |
| 144 | Pgk1 | 16119 | -0.570 | -0.0507 | No |
| 145 | Pdk1 | 16268 | -0.613 | -0.0413 | No |
| 146 | Maff | 16461 | -0.714 | -0.0315 | No |
| 147 | Rora | 16491 | -0.735 | -0.0113 | No |
| 148 | Selenbp1 | 16560 | -0.795 | 0.0084 | No |