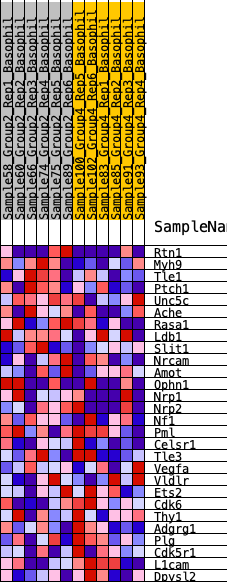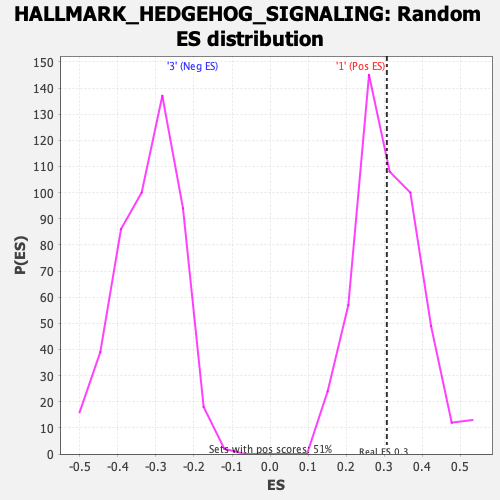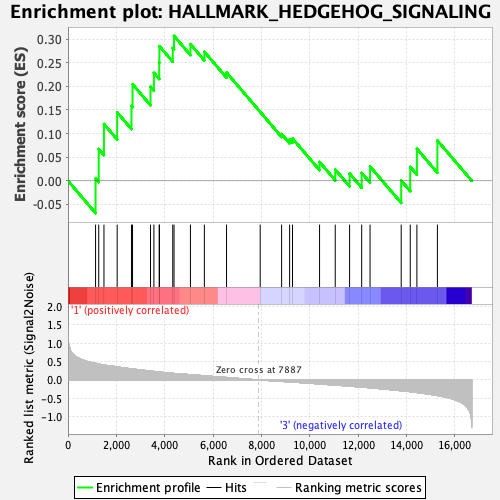
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.30682313 |
| Normalized Enrichment Score (NES) | 0.9904628 |
| Nominal p-value | 0.47047246 |
| FDR q-value | 0.6589354 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rtn1 | 1140 | 0.455 | 0.0049 | Yes |
| 2 | Myh9 | 1271 | 0.435 | 0.0673 | Yes |
| 3 | Tle1 | 1487 | 0.408 | 0.1202 | Yes |
| 4 | Ptch1 | 2033 | 0.356 | 0.1449 | Yes |
| 5 | Unc5c | 2625 | 0.304 | 0.1585 | Yes |
| 6 | Ache | 2669 | 0.300 | 0.2044 | Yes |
| 7 | Rasa1 | 3412 | 0.244 | 0.1992 | Yes |
| 8 | Ldb1 | 3552 | 0.234 | 0.2286 | Yes |
| 9 | Slit1 | 3772 | 0.217 | 0.2505 | Yes |
| 10 | Nrcam | 3780 | 0.217 | 0.2851 | Yes |
| 11 | Amot | 4332 | 0.181 | 0.2812 | Yes |
| 12 | Ophn1 | 4384 | 0.178 | 0.3068 | Yes |
| 13 | Nrp1 | 5063 | 0.144 | 0.2893 | No |
| 14 | Nrp2 | 5639 | 0.114 | 0.2732 | No |
| 15 | Nf1 | 6556 | 0.068 | 0.2293 | No |
| 16 | Pml | 7948 | -0.000 | 0.1459 | No |
| 17 | Celsr1 | 8834 | -0.039 | 0.0991 | No |
| 18 | Tle3 | 9165 | -0.051 | 0.0876 | No |
| 19 | Vegfa | 9284 | -0.057 | 0.0896 | No |
| 20 | Vldlr | 10403 | -0.108 | 0.0400 | No |
| 21 | Ets2 | 11051 | -0.141 | 0.0238 | No |
| 22 | Cdk6 | 11645 | -0.169 | 0.0155 | No |
| 23 | Thy1 | 12145 | -0.193 | 0.0167 | No |
| 24 | Adgrg1 | 12491 | -0.213 | 0.0304 | No |
| 25 | Plg | 13779 | -0.294 | 0.0006 | No |
| 26 | Cdk5r1 | 14153 | -0.318 | 0.0294 | No |
| 27 | L1cam | 14428 | -0.341 | 0.0679 | No |
| 28 | Dpysl2 | 15276 | -0.423 | 0.0854 | No |