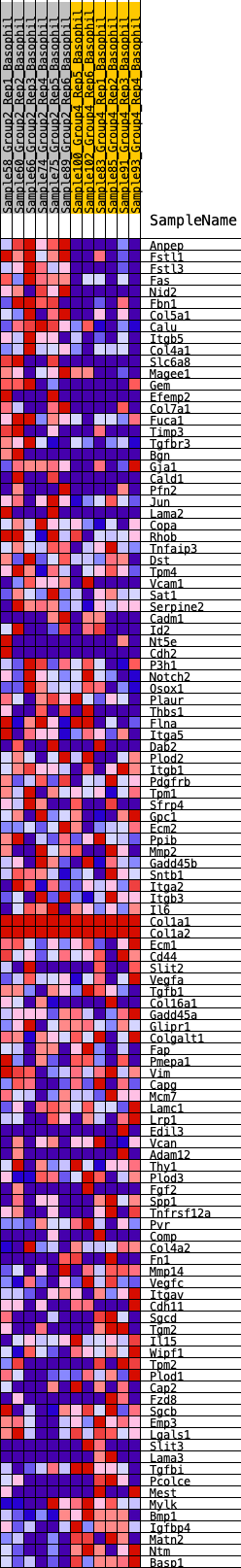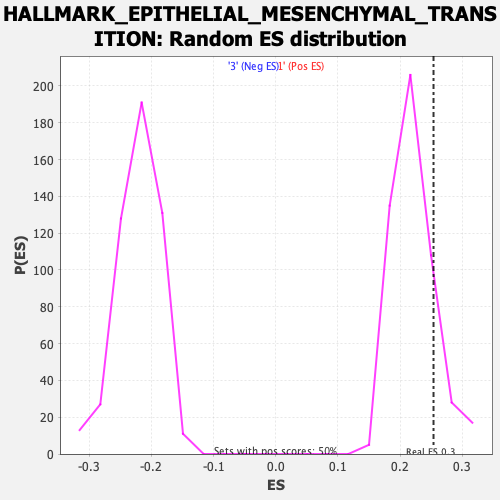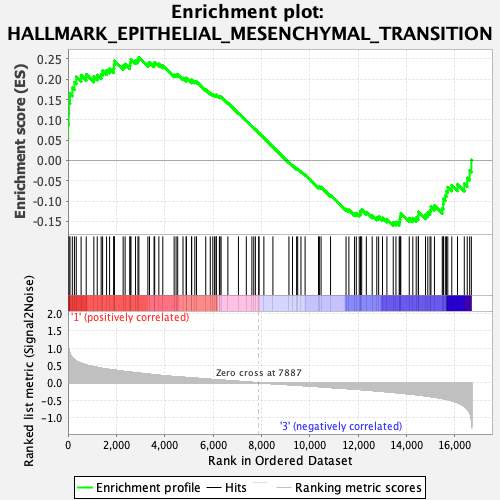
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2538831 |
| Normalized Enrichment Score (NES) | 1.1473105 |
| Nominal p-value | 0.1503006 |
| FDR q-value | 0.59806716 |
| FWER p-Value | 0.978 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Anpep | 0 | 1.911 | 0.0542 | Yes |
| 2 | Fstl1 | 4 | 1.240 | 0.0892 | Yes |
| 3 | Fstl3 | 28 | 0.992 | 0.1160 | Yes |
| 4 | Fas | 38 | 0.957 | 0.1426 | Yes |
| 5 | Nid2 | 82 | 0.864 | 0.1645 | Yes |
| 6 | Fbn1 | 179 | 0.722 | 0.1792 | Yes |
| 7 | Col5a1 | 266 | 0.667 | 0.1929 | Yes |
| 8 | Calu | 341 | 0.630 | 0.2064 | Yes |
| 9 | Itgb5 | 542 | 0.565 | 0.2103 | Yes |
| 10 | Col4a1 | 753 | 0.513 | 0.2122 | Yes |
| 11 | Slc6a8 | 1069 | 0.463 | 0.2064 | Yes |
| 12 | Magee1 | 1209 | 0.445 | 0.2106 | Yes |
| 13 | Gem | 1372 | 0.420 | 0.2128 | Yes |
| 14 | Efemp2 | 1431 | 0.414 | 0.2210 | Yes |
| 15 | Col7a1 | 1595 | 0.397 | 0.2224 | Yes |
| 16 | Fuca1 | 1719 | 0.385 | 0.2259 | Yes |
| 17 | Timp3 | 1889 | 0.371 | 0.2263 | Yes |
| 18 | Tgfbr3 | 1905 | 0.369 | 0.2358 | Yes |
| 19 | Bgn | 1919 | 0.368 | 0.2455 | Yes |
| 20 | Gja1 | 2280 | 0.332 | 0.2332 | Yes |
| 21 | Cald1 | 2362 | 0.326 | 0.2376 | Yes |
| 22 | Pfn2 | 2556 | 0.308 | 0.2347 | Yes |
| 23 | Jun | 2573 | 0.307 | 0.2424 | Yes |
| 24 | Lama2 | 2604 | 0.304 | 0.2493 | Yes |
| 25 | Copa | 2793 | 0.291 | 0.2462 | Yes |
| 26 | Rhob | 2893 | 0.282 | 0.2482 | Yes |
| 27 | Tnfaip3 | 2931 | 0.279 | 0.2539 | Yes |
| 28 | Dst | 3304 | 0.252 | 0.2386 | No |
| 29 | Tpm4 | 3370 | 0.247 | 0.2417 | No |
| 30 | Vcam1 | 3559 | 0.233 | 0.2370 | No |
| 31 | Sat1 | 3585 | 0.231 | 0.2420 | No |
| 32 | Serpine2 | 3757 | 0.219 | 0.2379 | No |
| 33 | Cadm1 | 3920 | 0.208 | 0.2340 | No |
| 34 | Id2 | 4389 | 0.178 | 0.2109 | No |
| 35 | Nt5e | 4470 | 0.173 | 0.2109 | No |
| 36 | Cdh2 | 4536 | 0.169 | 0.2118 | No |
| 37 | P3h1 | 4756 | 0.162 | 0.2032 | No |
| 38 | Notch2 | 4887 | 0.154 | 0.1997 | No |
| 39 | Qsox1 | 4894 | 0.153 | 0.2037 | No |
| 40 | Plaur | 5110 | 0.141 | 0.1948 | No |
| 41 | Thbs1 | 5113 | 0.141 | 0.1987 | No |
| 42 | Flna | 5244 | 0.134 | 0.1946 | No |
| 43 | Itga5 | 5315 | 0.130 | 0.1941 | No |
| 44 | Dab2 | 5696 | 0.111 | 0.1743 | No |
| 45 | Plod2 | 5883 | 0.101 | 0.1660 | No |
| 46 | Itgb1 | 5988 | 0.096 | 0.1625 | No |
| 47 | Pdgfrb | 6062 | 0.093 | 0.1607 | No |
| 48 | Tpm1 | 6126 | 0.090 | 0.1594 | No |
| 49 | Sfrp4 | 6132 | 0.089 | 0.1617 | No |
| 50 | Gpc1 | 6261 | 0.083 | 0.1563 | No |
| 51 | Ecm2 | 6285 | 0.081 | 0.1572 | No |
| 52 | Ppib | 6331 | 0.078 | 0.1567 | No |
| 53 | Mmp2 | 6611 | 0.065 | 0.1417 | No |
| 54 | Gadd45b | 7049 | 0.043 | 0.1166 | No |
| 55 | Sntb1 | 7374 | 0.025 | 0.0978 | No |
| 56 | Itga2 | 7605 | 0.014 | 0.0843 | No |
| 57 | Itgb3 | 7682 | 0.010 | 0.0800 | No |
| 58 | Il6 | 7749 | 0.007 | 0.0762 | No |
| 59 | Col1a1 | 7888 | 0.000 | 0.0679 | No |
| 60 | Col1a2 | 7899 | 0.000 | 0.0673 | No |
| 61 | Ecm1 | 8100 | -0.007 | 0.0554 | No |
| 62 | Cd44 | 8474 | -0.022 | 0.0335 | No |
| 63 | Slit2 | 9136 | -0.050 | -0.0049 | No |
| 64 | Vegfa | 9284 | -0.057 | -0.0122 | No |
| 65 | Tgfb1 | 9454 | -0.064 | -0.0205 | No |
| 66 | Col16a1 | 9498 | -0.066 | -0.0212 | No |
| 67 | Gadd45a | 9634 | -0.073 | -0.0273 | No |
| 68 | Glipr1 | 9805 | -0.081 | -0.0353 | No |
| 69 | Colgalt1 | 10359 | -0.106 | -0.0656 | No |
| 70 | Fap | 10396 | -0.108 | -0.0647 | No |
| 71 | Pmepa1 | 10466 | -0.111 | -0.0657 | No |
| 72 | Vim | 10857 | -0.131 | -0.0855 | No |
| 73 | Capg | 11498 | -0.160 | -0.1196 | No |
| 74 | Mcm7 | 11616 | -0.167 | -0.1219 | No |
| 75 | Lamc1 | 11849 | -0.178 | -0.1308 | No |
| 76 | Lrp1 | 11922 | -0.182 | -0.1300 | No |
| 77 | Edil3 | 12043 | -0.189 | -0.1319 | No |
| 78 | Vcan | 12089 | -0.190 | -0.1292 | No |
| 79 | Adam12 | 12101 | -0.191 | -0.1244 | No |
| 80 | Thy1 | 12145 | -0.193 | -0.1216 | No |
| 81 | Plod3 | 12337 | -0.203 | -0.1273 | No |
| 82 | Fgf2 | 12579 | -0.218 | -0.1356 | No |
| 83 | Spp1 | 12769 | -0.230 | -0.1405 | No |
| 84 | Tnfrsf12a | 12846 | -0.234 | -0.1385 | No |
| 85 | Pvr | 13006 | -0.244 | -0.1411 | No |
| 86 | Comp | 13188 | -0.255 | -0.1448 | No |
| 87 | Col4a2 | 13450 | -0.271 | -0.1529 | No |
| 88 | Fn1 | 13562 | -0.279 | -0.1516 | No |
| 89 | Mmp14 | 13696 | -0.288 | -0.1515 | No |
| 90 | Vegfc | 13723 | -0.290 | -0.1448 | No |
| 91 | Itgav | 13742 | -0.291 | -0.1376 | No |
| 92 | Cdh11 | 13762 | -0.293 | -0.1305 | No |
| 93 | Sgcd | 14112 | -0.314 | -0.1426 | No |
| 94 | Tgm2 | 14259 | -0.326 | -0.1422 | No |
| 95 | Il15 | 14399 | -0.338 | -0.1409 | No |
| 96 | Wipf1 | 14487 | -0.347 | -0.1363 | No |
| 97 | Tpm2 | 14489 | -0.347 | -0.1266 | No |
| 98 | Plod1 | 14784 | -0.372 | -0.1337 | No |
| 99 | Cap2 | 14879 | -0.380 | -0.1286 | No |
| 100 | Fzd8 | 14971 | -0.390 | -0.1230 | No |
| 101 | Sgcb | 15009 | -0.394 | -0.1141 | No |
| 102 | Emp3 | 15154 | -0.408 | -0.1112 | No |
| 103 | Lgals1 | 15477 | -0.448 | -0.1179 | No |
| 104 | Slit3 | 15516 | -0.452 | -0.1073 | No |
| 105 | Lama3 | 15522 | -0.452 | -0.0948 | No |
| 106 | Tgfbi | 15603 | -0.464 | -0.0865 | No |
| 107 | Pcolce | 15651 | -0.471 | -0.0760 | No |
| 108 | Mest | 15706 | -0.481 | -0.0656 | No |
| 109 | Mylk | 15871 | -0.513 | -0.0609 | No |
| 110 | Bmp1 | 16107 | -0.566 | -0.0590 | No |
| 111 | Igfbp4 | 16388 | -0.666 | -0.0570 | No |
| 112 | Matn2 | 16510 | -0.749 | -0.0431 | No |
| 113 | Ntm | 16611 | -0.857 | -0.0248 | No |
| 114 | Basp1 | 16680 | -1.059 | 0.0011 | No |