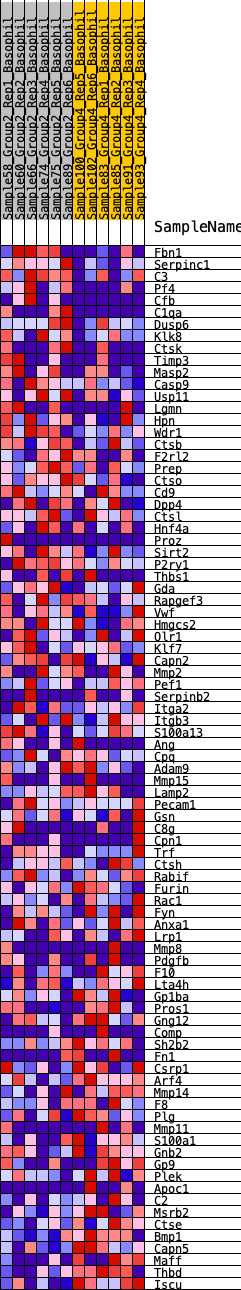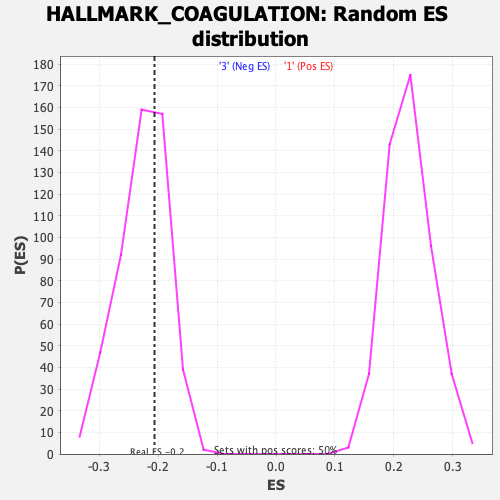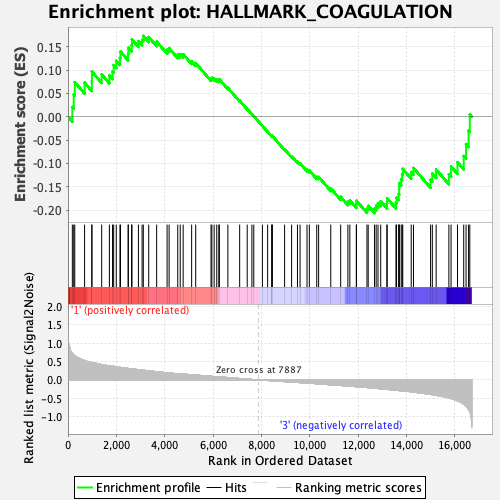
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.20634815 |
| Normalized Enrichment Score (NES) | -0.91584307 |
| Nominal p-value | 0.64484125 |
| FDR q-value | 0.9955218 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fbn1 | 179 | 0.722 | 0.0209 | No |
| 2 | Serpinc1 | 237 | 0.687 | 0.0476 | No |
| 3 | C3 | 279 | 0.662 | 0.0742 | No |
| 4 | Pf4 | 686 | 0.531 | 0.0730 | No |
| 5 | Cfb | 985 | 0.475 | 0.0759 | No |
| 6 | C1qa | 989 | 0.475 | 0.0966 | No |
| 7 | Dusp6 | 1393 | 0.418 | 0.0907 | No |
| 8 | Klk8 | 1711 | 0.385 | 0.0885 | No |
| 9 | Ctsk | 1842 | 0.375 | 0.0971 | No |
| 10 | Timp3 | 1889 | 0.371 | 0.1106 | No |
| 11 | Masp2 | 1996 | 0.361 | 0.1200 | No |
| 12 | Casp9 | 2149 | 0.344 | 0.1260 | No |
| 13 | Usp11 | 2170 | 0.342 | 0.1397 | No |
| 14 | Lgmn | 2487 | 0.314 | 0.1345 | No |
| 15 | Hpn | 2495 | 0.314 | 0.1478 | No |
| 16 | Wdr1 | 2629 | 0.304 | 0.1532 | No |
| 17 | Ctsb | 2645 | 0.303 | 0.1655 | No |
| 18 | F2rl2 | 2912 | 0.280 | 0.1618 | No |
| 19 | Prep | 3064 | 0.269 | 0.1645 | No |
| 20 | Ctso | 3114 | 0.265 | 0.1732 | No |
| 21 | Cd9 | 3338 | 0.249 | 0.1707 | No |
| 22 | Dpp4 | 3666 | 0.224 | 0.1609 | No |
| 23 | Ctsl | 4098 | 0.196 | 0.1435 | No |
| 24 | Hnf4a | 4181 | 0.191 | 0.1469 | No |
| 25 | Proz | 4539 | 0.169 | 0.1329 | No |
| 26 | Sirt2 | 4641 | 0.167 | 0.1341 | No |
| 27 | P2ry1 | 4762 | 0.161 | 0.1340 | No |
| 28 | Thbs1 | 5113 | 0.141 | 0.1191 | No |
| 29 | Gda | 5281 | 0.132 | 0.1149 | No |
| 30 | Rapgef3 | 5914 | 0.099 | 0.0812 | No |
| 31 | Vwf | 5950 | 0.098 | 0.0834 | No |
| 32 | Hmgcs2 | 6044 | 0.093 | 0.0819 | No |
| 33 | Olr1 | 6152 | 0.088 | 0.0793 | No |
| 34 | Klf7 | 6238 | 0.084 | 0.0778 | No |
| 35 | Capn2 | 6259 | 0.083 | 0.0803 | No |
| 36 | Mmp2 | 6611 | 0.065 | 0.0620 | No |
| 37 | Pef1 | 7099 | 0.040 | 0.0344 | No |
| 38 | Serpinb2 | 7411 | 0.023 | 0.0167 | No |
| 39 | Itga2 | 7605 | 0.014 | 0.0057 | No |
| 40 | Itgb3 | 7682 | 0.010 | 0.0016 | No |
| 41 | S100a13 | 8039 | -0.004 | -0.0197 | No |
| 42 | Ang | 8256 | -0.013 | -0.0321 | No |
| 43 | Cpq | 8426 | -0.020 | -0.0414 | No |
| 44 | Adam9 | 8433 | -0.020 | -0.0409 | No |
| 45 | Mmp15 | 8451 | -0.021 | -0.0410 | No |
| 46 | Lamp2 | 8957 | -0.043 | -0.0695 | No |
| 47 | Pecam1 | 9247 | -0.054 | -0.0845 | No |
| 48 | Gsn | 9489 | -0.066 | -0.0962 | No |
| 49 | C8g | 9596 | -0.071 | -0.0994 | No |
| 50 | Cpn1 | 9884 | -0.083 | -0.1131 | No |
| 51 | Trf | 9980 | -0.088 | -0.1149 | No |
| 52 | Ctsh | 10286 | -0.101 | -0.1288 | No |
| 53 | Rabif | 10364 | -0.106 | -0.1288 | No |
| 54 | Furin | 10866 | -0.131 | -0.1532 | No |
| 55 | Rac1 | 11276 | -0.150 | -0.1713 | No |
| 56 | Fyn | 11574 | -0.165 | -0.1819 | No |
| 57 | Anxa1 | 11655 | -0.169 | -0.1793 | No |
| 58 | Lrp1 | 11922 | -0.182 | -0.1873 | No |
| 59 | Mmp8 | 11931 | -0.183 | -0.1798 | No |
| 60 | Pdgfb | 12363 | -0.205 | -0.1967 | Yes |
| 61 | F10 | 12414 | -0.208 | -0.1906 | Yes |
| 62 | Lta4h | 12676 | -0.225 | -0.1965 | Yes |
| 63 | Gp1ba | 12743 | -0.228 | -0.1905 | Yes |
| 64 | Pros1 | 12820 | -0.232 | -0.1848 | Yes |
| 65 | Gng12 | 12932 | -0.238 | -0.1811 | Yes |
| 66 | Comp | 13188 | -0.255 | -0.1852 | Yes |
| 67 | Sh2b2 | 13197 | -0.255 | -0.1745 | Yes |
| 68 | Fn1 | 13562 | -0.279 | -0.1842 | Yes |
| 69 | Csrp1 | 13586 | -0.281 | -0.1733 | Yes |
| 70 | Arf4 | 13672 | -0.286 | -0.1658 | Yes |
| 71 | Mmp14 | 13696 | -0.288 | -0.1546 | Yes |
| 72 | F8 | 13697 | -0.288 | -0.1419 | Yes |
| 73 | Plg | 13779 | -0.294 | -0.1339 | Yes |
| 74 | Mmp11 | 13817 | -0.297 | -0.1231 | Yes |
| 75 | S100a1 | 13844 | -0.298 | -0.1116 | Yes |
| 76 | Gnb2 | 14193 | -0.321 | -0.1185 | Yes |
| 77 | Gp9 | 14286 | -0.329 | -0.1096 | Yes |
| 78 | Plek | 14994 | -0.392 | -0.1349 | Yes |
| 79 | Apoc1 | 15066 | -0.400 | -0.1217 | Yes |
| 80 | C2 | 15224 | -0.416 | -0.1129 | Yes |
| 81 | Msrb2 | 15751 | -0.487 | -0.1231 | Yes |
| 82 | Ctse | 15841 | -0.505 | -0.1063 | Yes |
| 83 | Bmp1 | 16107 | -0.566 | -0.0974 | Yes |
| 84 | Capn5 | 16364 | -0.655 | -0.0841 | Yes |
| 85 | Maff | 16461 | -0.714 | -0.0586 | Yes |
| 86 | Thbd | 16565 | -0.797 | -0.0298 | Yes |
| 87 | Iscu | 16616 | -0.862 | 0.0050 | Yes |