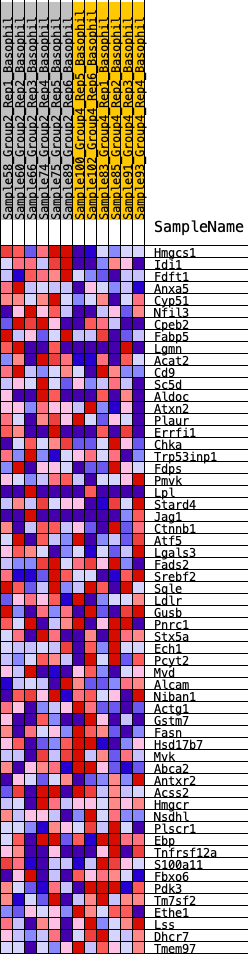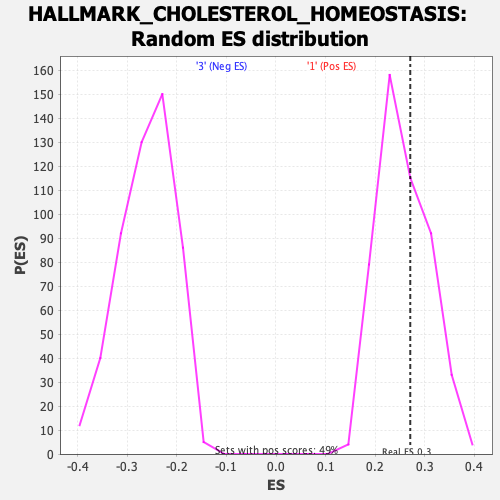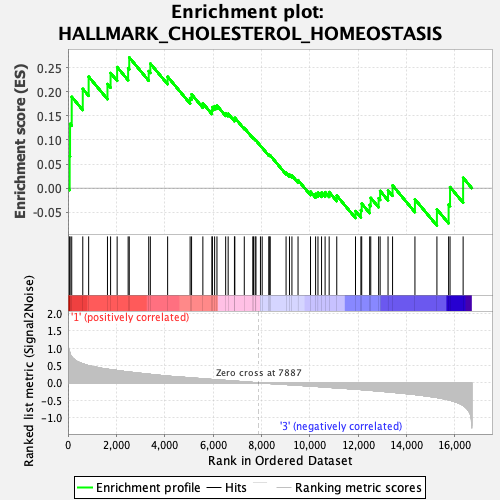
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.2710414 |
| Normalized Enrichment Score (NES) | 1.0466635 |
| Nominal p-value | 0.41030928 |
| FDR q-value | 0.6727722 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hmgcs1 | 69 | 0.884 | 0.0667 | Yes |
| 2 | Idi1 | 85 | 0.842 | 0.1333 | Yes |
| 3 | Fdft1 | 154 | 0.749 | 0.1893 | Yes |
| 4 | Anxa5 | 611 | 0.549 | 0.2059 | Yes |
| 5 | Cyp51 | 854 | 0.496 | 0.2311 | Yes |
| 6 | Nfil3 | 1632 | 0.392 | 0.2159 | Yes |
| 7 | Cpeb2 | 1761 | 0.380 | 0.2387 | Yes |
| 8 | Fabp5 | 2032 | 0.356 | 0.2510 | Yes |
| 9 | Lgmn | 2487 | 0.314 | 0.2489 | Yes |
| 10 | Acat2 | 2533 | 0.310 | 0.2710 | Yes |
| 11 | Cd9 | 3338 | 0.249 | 0.2427 | No |
| 12 | Sc5d | 3405 | 0.244 | 0.2583 | No |
| 13 | Aldoc | 4120 | 0.195 | 0.2310 | No |
| 14 | Atxn2 | 5050 | 0.144 | 0.1868 | No |
| 15 | Plaur | 5110 | 0.141 | 0.1946 | No |
| 16 | Errfi1 | 5577 | 0.117 | 0.1760 | No |
| 17 | Chka | 5951 | 0.098 | 0.1614 | No |
| 18 | Trp53inp1 | 5971 | 0.097 | 0.1680 | No |
| 19 | Fdps | 6063 | 0.093 | 0.1700 | No |
| 20 | Pmvk | 6160 | 0.088 | 0.1713 | No |
| 21 | Lpl | 6520 | 0.070 | 0.1553 | No |
| 22 | Stard4 | 6623 | 0.065 | 0.1543 | No |
| 23 | Jag1 | 6883 | 0.050 | 0.1428 | No |
| 24 | Ctnnb1 | 6899 | 0.049 | 0.1458 | No |
| 25 | Atf5 | 7290 | 0.029 | 0.1248 | No |
| 26 | Lgals3 | 7642 | 0.012 | 0.1046 | No |
| 27 | Fads2 | 7673 | 0.010 | 0.1037 | No |
| 28 | Srebf2 | 7740 | 0.007 | 0.1002 | No |
| 29 | Sqle | 7773 | 0.006 | 0.0988 | No |
| 30 | Ldlr | 7958 | -0.001 | 0.0878 | No |
| 31 | Gusb | 8030 | -0.004 | 0.0838 | No |
| 32 | Pnrc1 | 8307 | -0.015 | 0.0684 | No |
| 33 | Stx5a | 8321 | -0.016 | 0.0689 | No |
| 34 | Ech1 | 8363 | -0.017 | 0.0678 | No |
| 35 | Pcyt2 | 9018 | -0.045 | 0.0321 | No |
| 36 | Mvd | 9159 | -0.051 | 0.0278 | No |
| 37 | Alcam | 9260 | -0.055 | 0.0262 | No |
| 38 | Niban1 | 9513 | -0.067 | 0.0165 | No |
| 39 | Actg1 | 10029 | -0.090 | -0.0073 | No |
| 40 | Gstm7 | 10239 | -0.099 | -0.0119 | No |
| 41 | Fasn | 10337 | -0.104 | -0.0094 | No |
| 42 | Hsd17b7 | 10485 | -0.112 | -0.0092 | No |
| 43 | Mvk | 10632 | -0.120 | -0.0084 | No |
| 44 | Abca2 | 10800 | -0.128 | -0.0082 | No |
| 45 | Antxr2 | 11114 | -0.143 | -0.0155 | No |
| 46 | Acss2 | 11889 | -0.180 | -0.0476 | No |
| 47 | Hmgcr | 12113 | -0.191 | -0.0457 | No |
| 48 | Nsdhl | 12146 | -0.193 | -0.0321 | No |
| 49 | Plscr1 | 12475 | -0.212 | -0.0348 | No |
| 50 | Ebp | 12520 | -0.215 | -0.0203 | No |
| 51 | Tnfrsf12a | 12846 | -0.234 | -0.0211 | No |
| 52 | S100a11 | 12908 | -0.237 | -0.0058 | No |
| 53 | Fbxo6 | 13235 | -0.257 | -0.0048 | No |
| 54 | Pdk3 | 13422 | -0.269 | 0.0056 | No |
| 55 | Tm7sf2 | 14347 | -0.334 | -0.0231 | No |
| 56 | Ethe1 | 15256 | -0.420 | -0.0440 | No |
| 57 | Lss | 15740 | -0.486 | -0.0341 | No |
| 58 | Dhcr7 | 15799 | -0.496 | 0.0022 | No |
| 59 | Tmem97 | 16340 | -0.647 | 0.0216 | No |