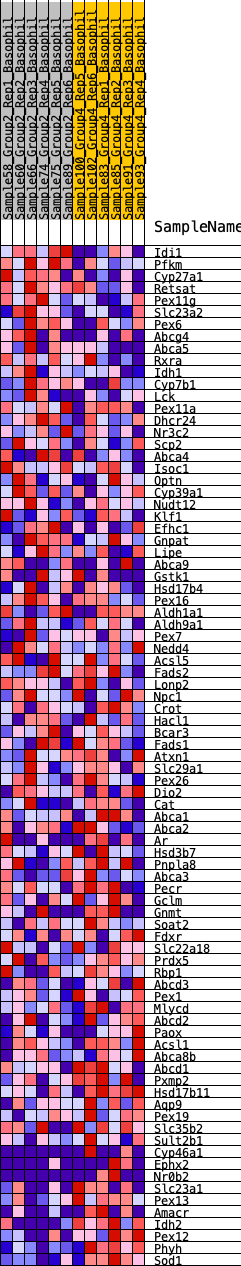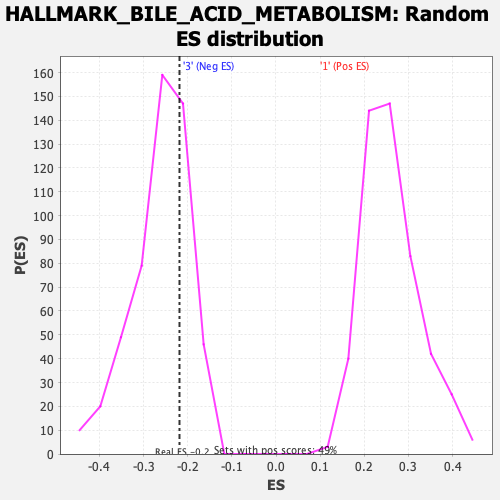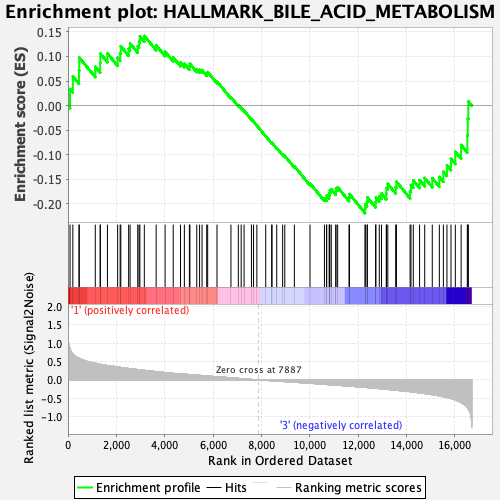
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.21859936 |
| Normalized Enrichment Score (NES) | -0.831174 |
| Nominal p-value | 0.7372549 |
| FDR q-value | 0.9853644 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idi1 | 85 | 0.842 | 0.0336 | No |
| 2 | Pfkm | 200 | 0.708 | 0.0594 | No |
| 3 | Cyp27a1 | 454 | 0.591 | 0.0714 | No |
| 4 | Retsat | 466 | 0.586 | 0.0977 | No |
| 5 | Pex11g | 1128 | 0.456 | 0.0789 | No |
| 6 | Slc23a2 | 1330 | 0.427 | 0.0865 | No |
| 7 | Pex6 | 1337 | 0.426 | 0.1057 | No |
| 8 | Abcg4 | 1631 | 0.393 | 0.1061 | No |
| 9 | Abca5 | 2052 | 0.353 | 0.0971 | No |
| 10 | Rxra | 2156 | 0.343 | 0.1067 | No |
| 11 | Idh1 | 2184 | 0.341 | 0.1208 | No |
| 12 | Cyp7b1 | 2508 | 0.313 | 0.1158 | No |
| 13 | Lck | 2568 | 0.307 | 0.1264 | No |
| 14 | Pex11a | 2884 | 0.283 | 0.1204 | No |
| 15 | Dhcr24 | 2938 | 0.278 | 0.1301 | No |
| 16 | Nr3c2 | 2972 | 0.275 | 0.1407 | No |
| 17 | Scp2 | 3156 | 0.262 | 0.1418 | No |
| 18 | Abca4 | 3645 | 0.226 | 0.1228 | No |
| 19 | Isoc1 | 4015 | 0.201 | 0.1098 | No |
| 20 | Optn | 4351 | 0.180 | 0.0980 | No |
| 21 | Cyp39a1 | 4652 | 0.167 | 0.0876 | No |
| 22 | Nudt12 | 4812 | 0.158 | 0.0853 | No |
| 23 | Klf1 | 5019 | 0.146 | 0.0796 | No |
| 24 | Efhc1 | 5042 | 0.145 | 0.0850 | No |
| 25 | Gnpat | 5325 | 0.129 | 0.0739 | No |
| 26 | Lipe | 5441 | 0.125 | 0.0728 | No |
| 27 | Abca9 | 5543 | 0.120 | 0.0722 | No |
| 28 | Gstk1 | 5736 | 0.109 | 0.0657 | No |
| 29 | Hsd17b4 | 5776 | 0.107 | 0.0683 | No |
| 30 | Pex16 | 6161 | 0.088 | 0.0492 | No |
| 31 | Aldh1a1 | 6738 | 0.058 | 0.0172 | No |
| 32 | Aldh9a1 | 7043 | 0.043 | 0.0009 | No |
| 33 | Pex7 | 7161 | 0.037 | -0.0045 | No |
| 34 | Nedd4 | 7281 | 0.030 | -0.0103 | No |
| 35 | Acsl5 | 7586 | 0.015 | -0.0278 | No |
| 36 | Fads2 | 7673 | 0.010 | -0.0325 | No |
| 37 | Lonp2 | 7810 | 0.004 | -0.0406 | No |
| 38 | Npc1 | 8176 | -0.010 | -0.0621 | No |
| 39 | Crot | 8424 | -0.020 | -0.0760 | No |
| 40 | Hacl1 | 8436 | -0.020 | -0.0758 | No |
| 41 | Bcar3 | 8631 | -0.029 | -0.0861 | No |
| 42 | Fads1 | 8876 | -0.040 | -0.0990 | No |
| 43 | Atxn1 | 8965 | -0.043 | -0.1023 | No |
| 44 | Slc29a1 | 9361 | -0.060 | -0.1233 | No |
| 45 | Pex26 | 10008 | -0.089 | -0.1581 | No |
| 46 | Dio2 | 10605 | -0.118 | -0.1885 | No |
| 47 | Cat | 10693 | -0.122 | -0.1881 | No |
| 48 | Abca1 | 10697 | -0.122 | -0.1826 | No |
| 49 | Abca2 | 10800 | -0.128 | -0.1829 | No |
| 50 | Ar | 10814 | -0.129 | -0.1778 | No |
| 51 | Hsd3b7 | 10826 | -0.129 | -0.1725 | No |
| 52 | Pnpla8 | 10887 | -0.132 | -0.1700 | No |
| 53 | Abca3 | 11075 | -0.142 | -0.1747 | No |
| 54 | Pecr | 11080 | -0.142 | -0.1684 | No |
| 55 | Gclm | 11151 | -0.145 | -0.1659 | No |
| 56 | Gnmt | 11621 | -0.167 | -0.1865 | No |
| 57 | Soat2 | 11641 | -0.169 | -0.1798 | No |
| 58 | Fdxr | 12286 | -0.201 | -0.2093 | Yes |
| 59 | Slc22a18 | 12291 | -0.201 | -0.2003 | Yes |
| 60 | Prdx5 | 12367 | -0.205 | -0.1954 | Yes |
| 61 | Rbp1 | 12381 | -0.206 | -0.1867 | Yes |
| 62 | Abcd3 | 12720 | -0.227 | -0.1965 | Yes |
| 63 | Pex1 | 12733 | -0.228 | -0.1868 | Yes |
| 64 | Mlycd | 12873 | -0.235 | -0.1843 | Yes |
| 65 | Abcd2 | 12964 | -0.241 | -0.1787 | Yes |
| 66 | Paox | 13164 | -0.254 | -0.1790 | Yes |
| 67 | Acsl1 | 13168 | -0.254 | -0.1675 | Yes |
| 68 | Abca8b | 13225 | -0.256 | -0.1590 | Yes |
| 69 | Abcd1 | 13551 | -0.278 | -0.1658 | Yes |
| 70 | Pxmp2 | 13581 | -0.280 | -0.1546 | Yes |
| 71 | Hsd17b11 | 14147 | -0.317 | -0.1740 | Yes |
| 72 | Aqp9 | 14183 | -0.320 | -0.1614 | Yes |
| 73 | Pex19 | 14279 | -0.328 | -0.1520 | Yes |
| 74 | Slc35b2 | 14539 | -0.351 | -0.1514 | Yes |
| 75 | Sult2b1 | 14751 | -0.369 | -0.1471 | Yes |
| 76 | Cyp46a1 | 15064 | -0.400 | -0.1474 | Yes |
| 77 | Ephx2 | 15356 | -0.433 | -0.1450 | Yes |
| 78 | Nr0b2 | 15523 | -0.452 | -0.1342 | Yes |
| 79 | Slc23a1 | 15671 | -0.473 | -0.1213 | Yes |
| 80 | Pex13 | 15838 | -0.504 | -0.1080 | Yes |
| 81 | Amacr | 16017 | -0.546 | -0.0936 | Yes |
| 82 | Idh2 | 16257 | -0.608 | -0.0800 | Yes |
| 83 | Pex12 | 16513 | -0.751 | -0.0608 | Yes |
| 84 | Phyh | 16533 | -0.769 | -0.0265 | Yes |
| 85 | Sod1 | 16556 | -0.791 | 0.0086 | Yes |