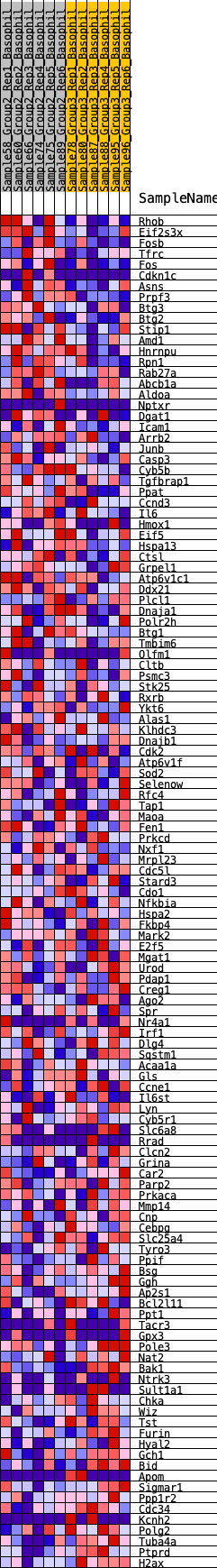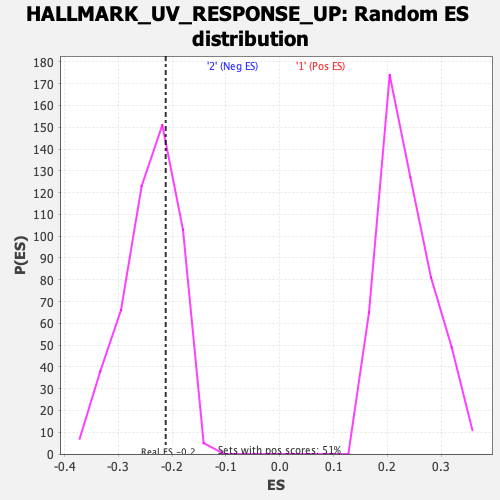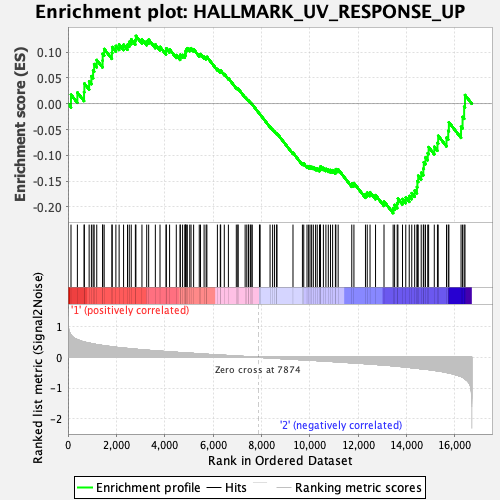
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.21213417 |
| Normalized Enrichment Score (NES) | -0.876539 |
| Nominal p-value | 0.7119675 |
| FDR q-value | 0.9007633 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rhob | 124 | 0.720 | 0.0177 | No |
| 2 | Eif2s3x | 388 | 0.571 | 0.0219 | No |
| 3 | Fosb | 664 | 0.498 | 0.0227 | No |
| 4 | Tfrc | 677 | 0.495 | 0.0393 | No |
| 5 | Fos | 874 | 0.462 | 0.0437 | No |
| 6 | Cdkn1c | 968 | 0.443 | 0.0536 | No |
| 7 | Asns | 1038 | 0.433 | 0.0646 | No |
| 8 | Prpf3 | 1081 | 0.427 | 0.0770 | No |
| 9 | Btg3 | 1189 | 0.411 | 0.0850 | No |
| 10 | Btg2 | 1430 | 0.381 | 0.0838 | No |
| 11 | Stip1 | 1435 | 0.380 | 0.0969 | No |
| 12 | Amd1 | 1496 | 0.372 | 0.1063 | No |
| 13 | Hnrnpu | 1816 | 0.338 | 0.0989 | No |
| 14 | Rpn1 | 1827 | 0.336 | 0.1101 | No |
| 15 | Rab27a | 1980 | 0.324 | 0.1123 | No |
| 16 | Abcb1a | 2114 | 0.312 | 0.1152 | No |
| 17 | Aldoa | 2298 | 0.298 | 0.1146 | No |
| 18 | Nptxr | 2461 | 0.286 | 0.1148 | No |
| 19 | Dgat1 | 2533 | 0.279 | 0.1204 | No |
| 20 | Icam1 | 2615 | 0.272 | 0.1250 | No |
| 21 | Arrb2 | 2786 | 0.261 | 0.1239 | No |
| 22 | Junb | 2808 | 0.260 | 0.1317 | No |
| 23 | Casp3 | 3058 | 0.241 | 0.1252 | No |
| 24 | Cyb5b | 3255 | 0.230 | 0.1214 | No |
| 25 | Tgfbrap1 | 3336 | 0.225 | 0.1244 | No |
| 26 | Ppat | 3612 | 0.207 | 0.1151 | No |
| 27 | Ccnd3 | 3805 | 0.197 | 0.1104 | No |
| 28 | Il6 | 4054 | 0.182 | 0.1018 | No |
| 29 | Hmox1 | 4066 | 0.182 | 0.1075 | No |
| 30 | Eif5 | 4201 | 0.174 | 0.1055 | No |
| 31 | Hspa13 | 4478 | 0.161 | 0.0945 | No |
| 32 | Ctsl | 4636 | 0.153 | 0.0904 | No |
| 33 | Grpel1 | 4643 | 0.152 | 0.0954 | No |
| 34 | Atp6v1c1 | 4739 | 0.146 | 0.0948 | No |
| 35 | Ddx21 | 4831 | 0.142 | 0.0943 | No |
| 36 | Plcl1 | 4855 | 0.141 | 0.0978 | No |
| 37 | Dnaja1 | 4864 | 0.140 | 0.1022 | No |
| 38 | Polr2h | 4890 | 0.139 | 0.1056 | No |
| 39 | Btg1 | 4934 | 0.137 | 0.1078 | No |
| 40 | Tmbim6 | 5035 | 0.132 | 0.1064 | No |
| 41 | Olfm1 | 5088 | 0.130 | 0.1078 | No |
| 42 | Cltb | 5195 | 0.125 | 0.1058 | No |
| 43 | Psmc3 | 5428 | 0.112 | 0.0957 | No |
| 44 | Stk25 | 5479 | 0.109 | 0.0965 | No |
| 45 | Rxrb | 5628 | 0.101 | 0.0911 | No |
| 46 | Ykt6 | 5711 | 0.097 | 0.0896 | No |
| 47 | Alas1 | 5743 | 0.096 | 0.0911 | No |
| 48 | Klhdc3 | 6178 | 0.076 | 0.0676 | No |
| 49 | Dnajb1 | 6298 | 0.071 | 0.0629 | No |
| 50 | Cdk2 | 6313 | 0.071 | 0.0645 | No |
| 51 | Atp6v1f | 6463 | 0.063 | 0.0578 | No |
| 52 | Sod2 | 6633 | 0.055 | 0.0495 | No |
| 53 | Selenow | 6960 | 0.040 | 0.0312 | No |
| 54 | Rfc4 | 7026 | 0.037 | 0.0286 | No |
| 55 | Tap1 | 7032 | 0.037 | 0.0296 | No |
| 56 | Maoa | 7325 | 0.024 | 0.0128 | No |
| 57 | Fen1 | 7393 | 0.021 | 0.0095 | No |
| 58 | Prkcd | 7460 | 0.018 | 0.0061 | No |
| 59 | Nxf1 | 7472 | 0.017 | 0.0061 | No |
| 60 | Mrpl23 | 7542 | 0.015 | 0.0024 | No |
| 61 | Cdc5l | 7573 | 0.013 | 0.0011 | No |
| 62 | Stard3 | 7620 | 0.011 | -0.0013 | No |
| 63 | Cdo1 | 7918 | -0.001 | -0.0192 | No |
| 64 | Nfkbia | 7945 | -0.002 | -0.0207 | No |
| 65 | Hspa2 | 8356 | -0.021 | -0.0447 | No |
| 66 | Fkbp4 | 8462 | -0.026 | -0.0502 | No |
| 67 | Mark2 | 8542 | -0.029 | -0.0539 | No |
| 68 | E2f5 | 8629 | -0.034 | -0.0579 | No |
| 69 | Mgat1 | 8640 | -0.034 | -0.0573 | No |
| 70 | Urod | 9304 | -0.064 | -0.0951 | No |
| 71 | Pdap1 | 9694 | -0.081 | -0.1157 | No |
| 72 | Creg1 | 9749 | -0.083 | -0.1160 | No |
| 73 | Ago2 | 9884 | -0.089 | -0.1210 | No |
| 74 | Spr | 9951 | -0.092 | -0.1217 | No |
| 75 | Nr4a1 | 10008 | -0.095 | -0.1218 | No |
| 76 | Irf1 | 10075 | -0.097 | -0.1224 | No |
| 77 | Dlg4 | 10145 | -0.100 | -0.1231 | No |
| 78 | Sqstm1 | 10238 | -0.104 | -0.1250 | No |
| 79 | Acaa1a | 10306 | -0.107 | -0.1253 | No |
| 80 | Gls | 10400 | -0.112 | -0.1269 | No |
| 81 | Ccne1 | 10434 | -0.113 | -0.1250 | No |
| 82 | Il6st | 10442 | -0.113 | -0.1214 | No |
| 83 | Lyn | 10561 | -0.120 | -0.1243 | No |
| 84 | Cyb5r1 | 10665 | -0.124 | -0.1262 | No |
| 85 | Slc6a8 | 10764 | -0.130 | -0.1276 | No |
| 86 | Rrad | 10855 | -0.133 | -0.1284 | No |
| 87 | Clcn2 | 10943 | -0.137 | -0.1288 | No |
| 88 | Grina | 11057 | -0.143 | -0.1306 | No |
| 89 | Car2 | 11077 | -0.144 | -0.1267 | No |
| 90 | Parp2 | 11172 | -0.148 | -0.1272 | No |
| 91 | Prkaca | 11735 | -0.179 | -0.1548 | No |
| 92 | Mmp14 | 11824 | -0.182 | -0.1538 | No |
| 93 | Cnp | 12301 | -0.207 | -0.1753 | No |
| 94 | Cebpg | 12374 | -0.211 | -0.1722 | No |
| 95 | Slc25a4 | 12490 | -0.217 | -0.1715 | No |
| 96 | Tyro3 | 12718 | -0.230 | -0.1772 | No |
| 97 | Ppif | 13068 | -0.252 | -0.1894 | No |
| 98 | Bsg | 13446 | -0.277 | -0.2024 | Yes |
| 99 | Ggh | 13506 | -0.281 | -0.1961 | Yes |
| 100 | Ap2s1 | 13614 | -0.289 | -0.1925 | Yes |
| 101 | Bcl2l11 | 13643 | -0.291 | -0.1840 | Yes |
| 102 | Ppt1 | 13833 | -0.305 | -0.1847 | Yes |
| 103 | Tacr3 | 13968 | -0.315 | -0.1818 | Yes |
| 104 | Gpx3 | 14111 | -0.328 | -0.1788 | Yes |
| 105 | Pole3 | 14216 | -0.338 | -0.1733 | Yes |
| 106 | Nat2 | 14336 | -0.348 | -0.1682 | Yes |
| 107 | Bak1 | 14430 | -0.355 | -0.1614 | Yes |
| 108 | Ntrk3 | 14448 | -0.357 | -0.1499 | Yes |
| 109 | Sult1a1 | 14482 | -0.360 | -0.1393 | Yes |
| 110 | Chka | 14606 | -0.372 | -0.1336 | Yes |
| 111 | Wiz | 14694 | -0.380 | -0.1256 | Yes |
| 112 | Tst | 14714 | -0.381 | -0.1134 | Yes |
| 113 | Furin | 14782 | -0.386 | -0.1039 | Yes |
| 114 | Hyal2 | 14877 | -0.394 | -0.0958 | Yes |
| 115 | Gch1 | 14913 | -0.397 | -0.0840 | Yes |
| 116 | Bid | 15147 | -0.423 | -0.0832 | Yes |
| 117 | Apom | 15278 | -0.438 | -0.0757 | Yes |
| 118 | Sigmar1 | 15308 | -0.442 | -0.0620 | Yes |
| 119 | Ppp1r2 | 15653 | -0.491 | -0.0655 | Yes |
| 120 | Cdc34 | 15724 | -0.504 | -0.0521 | Yes |
| 121 | Kcnh2 | 15751 | -0.508 | -0.0359 | Yes |
| 122 | Polg2 | 16254 | -0.621 | -0.0444 | Yes |
| 123 | Tuba4a | 16317 | -0.646 | -0.0255 | Yes |
| 124 | Ptprd | 16382 | -0.681 | -0.0055 | Yes |
| 125 | H2ax | 16417 | -0.700 | 0.0170 | Yes |