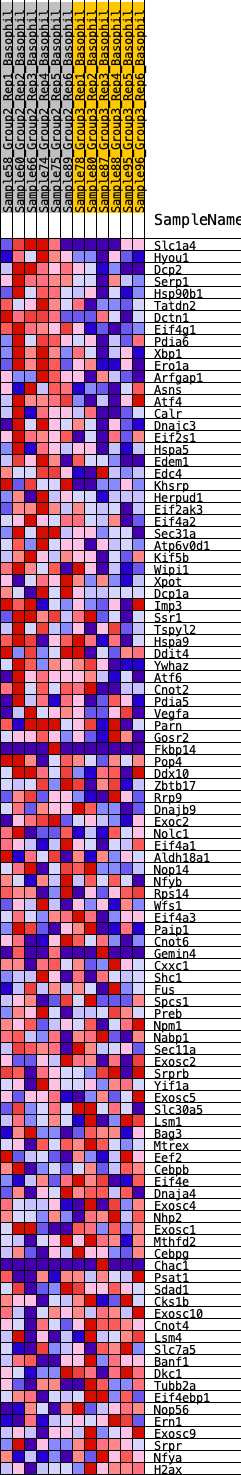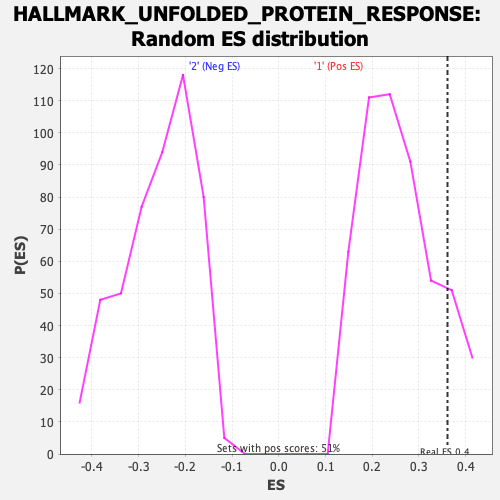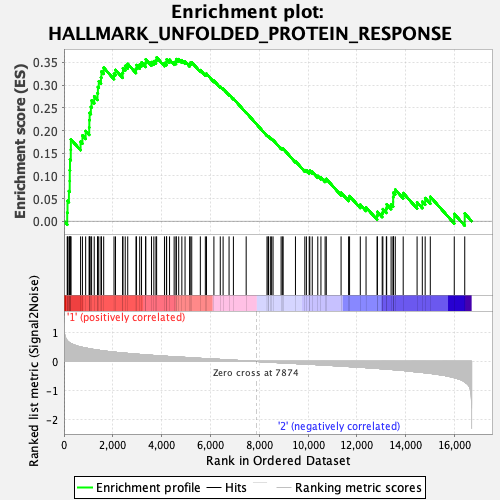
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.3611571 |
| Normalized Enrichment Score (NES) | 1.404343 |
| Nominal p-value | 0.125 |
| FDR q-value | 0.7973234 |
| FWER p-Value | 0.63 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc1a4 | 132 | 0.704 | 0.0190 | Yes |
| 2 | Hyou1 | 144 | 0.696 | 0.0449 | Yes |
| 3 | Dcp2 | 201 | 0.654 | 0.0665 | Yes |
| 4 | Serp1 | 234 | 0.630 | 0.0886 | Yes |
| 5 | Hsp90b1 | 237 | 0.627 | 0.1124 | Yes |
| 6 | Tatdn2 | 246 | 0.625 | 0.1358 | Yes |
| 7 | Dctn1 | 278 | 0.611 | 0.1573 | Yes |
| 8 | Eif4g1 | 282 | 0.609 | 0.1804 | Yes |
| 9 | Pdia6 | 679 | 0.494 | 0.1754 | Yes |
| 10 | Xbp1 | 755 | 0.480 | 0.1892 | Yes |
| 11 | Ero1a | 887 | 0.460 | 0.1989 | Yes |
| 12 | Arfgap1 | 1033 | 0.434 | 0.2067 | Yes |
| 13 | Asns | 1038 | 0.433 | 0.2230 | Yes |
| 14 | Atf4 | 1051 | 0.431 | 0.2388 | Yes |
| 15 | Calr | 1101 | 0.424 | 0.2520 | Yes |
| 16 | Dnajc3 | 1131 | 0.420 | 0.2663 | Yes |
| 17 | Eif2s1 | 1238 | 0.404 | 0.2754 | Yes |
| 18 | Hspa5 | 1372 | 0.388 | 0.2822 | Yes |
| 19 | Edem1 | 1389 | 0.386 | 0.2960 | Yes |
| 20 | Edc4 | 1426 | 0.381 | 0.3084 | Yes |
| 21 | Khsrp | 1515 | 0.371 | 0.3172 | Yes |
| 22 | Herpud1 | 1534 | 0.369 | 0.3302 | Yes |
| 23 | Eif2ak3 | 1626 | 0.361 | 0.3385 | Yes |
| 24 | Eif4a2 | 2049 | 0.318 | 0.3253 | Yes |
| 25 | Sec31a | 2110 | 0.312 | 0.3336 | Yes |
| 26 | Atp6v0d1 | 2401 | 0.290 | 0.3272 | Yes |
| 27 | Kif5b | 2415 | 0.289 | 0.3374 | Yes |
| 28 | Wipi1 | 2508 | 0.281 | 0.3426 | Yes |
| 29 | Xpot | 2611 | 0.273 | 0.3469 | Yes |
| 30 | Dcp1a | 2949 | 0.249 | 0.3361 | Yes |
| 31 | Imp3 | 2971 | 0.248 | 0.3443 | Yes |
| 32 | Ssr1 | 3102 | 0.239 | 0.3456 | Yes |
| 33 | Tspyl2 | 3176 | 0.235 | 0.3501 | Yes |
| 34 | Hspa9 | 3344 | 0.224 | 0.3486 | Yes |
| 35 | Ddit4 | 3349 | 0.224 | 0.3570 | Yes |
| 36 | Ywhaz | 3584 | 0.208 | 0.3508 | Yes |
| 37 | Atf6 | 3681 | 0.203 | 0.3528 | Yes |
| 38 | Cnot2 | 3767 | 0.199 | 0.3552 | Yes |
| 39 | Pdia5 | 3795 | 0.197 | 0.3612 | Yes |
| 40 | Vegfa | 4116 | 0.179 | 0.3487 | No |
| 41 | Parn | 4196 | 0.174 | 0.3506 | No |
| 42 | Gosr2 | 4205 | 0.174 | 0.3568 | No |
| 43 | Fkbp14 | 4324 | 0.168 | 0.3561 | No |
| 44 | Pop4 | 4512 | 0.159 | 0.3509 | No |
| 45 | Ddx10 | 4589 | 0.155 | 0.3522 | No |
| 46 | Zbtb17 | 4601 | 0.154 | 0.3574 | No |
| 47 | Rrp9 | 4699 | 0.149 | 0.3573 | No |
| 48 | Dnajb9 | 4828 | 0.142 | 0.3550 | No |
| 49 | Exoc2 | 4959 | 0.136 | 0.3523 | No |
| 50 | Nolc1 | 5146 | 0.127 | 0.3460 | No |
| 51 | Eif4a1 | 5170 | 0.126 | 0.3494 | No |
| 52 | Aldh18a1 | 5233 | 0.122 | 0.3504 | No |
| 53 | Nop14 | 5585 | 0.103 | 0.3332 | No |
| 54 | Nfyb | 5790 | 0.094 | 0.3244 | No |
| 55 | Rps14 | 5837 | 0.091 | 0.3252 | No |
| 56 | Wfs1 | 6141 | 0.078 | 0.3099 | No |
| 57 | Eif4a3 | 6402 | 0.066 | 0.2967 | No |
| 58 | Paip1 | 6518 | 0.061 | 0.2921 | No |
| 59 | Cnot6 | 6762 | 0.049 | 0.2794 | No |
| 60 | Gemin4 | 6940 | 0.041 | 0.2703 | No |
| 61 | Cxxc1 | 7463 | 0.018 | 0.2395 | No |
| 62 | Shc1 | 8316 | -0.019 | 0.1889 | No |
| 63 | Fus | 8359 | -0.021 | 0.1872 | No |
| 64 | Spcs1 | 8377 | -0.022 | 0.1870 | No |
| 65 | Preb | 8453 | -0.025 | 0.1834 | No |
| 66 | Npm1 | 8506 | -0.028 | 0.1814 | No |
| 67 | Nabp1 | 8564 | -0.030 | 0.1791 | No |
| 68 | Sec11a | 8894 | -0.046 | 0.1610 | No |
| 69 | Exosc2 | 8944 | -0.048 | 0.1599 | No |
| 70 | Srprb | 8978 | -0.050 | 0.1598 | No |
| 71 | Yif1a | 9482 | -0.071 | 0.1323 | No |
| 72 | Exosc5 | 9866 | -0.088 | 0.1126 | No |
| 73 | Slc30a5 | 9931 | -0.091 | 0.1122 | No |
| 74 | Lsm1 | 10046 | -0.096 | 0.1090 | No |
| 75 | Bag3 | 10070 | -0.097 | 0.1113 | No |
| 76 | Mtrex | 10170 | -0.101 | 0.1092 | No |
| 77 | Eef2 | 10397 | -0.112 | 0.0999 | No |
| 78 | Cebpb | 10526 | -0.118 | 0.0967 | No |
| 79 | Eif4e | 10697 | -0.126 | 0.0912 | No |
| 80 | Dnaja4 | 10745 | -0.128 | 0.0933 | No |
| 81 | Exosc4 | 11351 | -0.157 | 0.0628 | No |
| 82 | Nhp2 | 11659 | -0.174 | 0.0510 | No |
| 83 | Exosc1 | 11699 | -0.177 | 0.0554 | No |
| 84 | Mthfd2 | 12138 | -0.199 | 0.0366 | No |
| 85 | Cebpg | 12374 | -0.211 | 0.0305 | No |
| 86 | Chac1 | 12827 | -0.238 | 0.0123 | No |
| 87 | Psat1 | 12840 | -0.238 | 0.0207 | No |
| 88 | Sdad1 | 13036 | -0.250 | 0.0185 | No |
| 89 | Cks1b | 13063 | -0.252 | 0.0266 | No |
| 90 | Exosc10 | 13205 | -0.261 | 0.0280 | No |
| 91 | Cnot4 | 13219 | -0.262 | 0.0372 | No |
| 92 | Lsm4 | 13400 | -0.273 | 0.0368 | No |
| 93 | Slc7a5 | 13484 | -0.279 | 0.0425 | No |
| 94 | Banf1 | 13485 | -0.279 | 0.0532 | No |
| 95 | Dkc1 | 13497 | -0.280 | 0.0632 | No |
| 96 | Tubb2a | 13571 | -0.286 | 0.0697 | No |
| 97 | Eif4ebp1 | 13896 | -0.310 | 0.0621 | No |
| 98 | Nop56 | 14465 | -0.359 | 0.0415 | No |
| 99 | Ern1 | 14679 | -0.379 | 0.0432 | No |
| 100 | Exosc9 | 14796 | -0.386 | 0.0509 | No |
| 101 | Srpr | 15006 | -0.408 | 0.0539 | No |
| 102 | Nfya | 15988 | -0.555 | 0.0160 | No |
| 103 | H2ax | 16417 | -0.700 | 0.0170 | No |