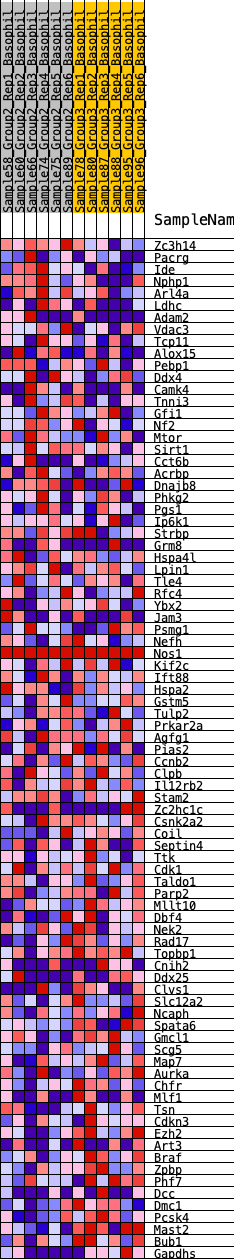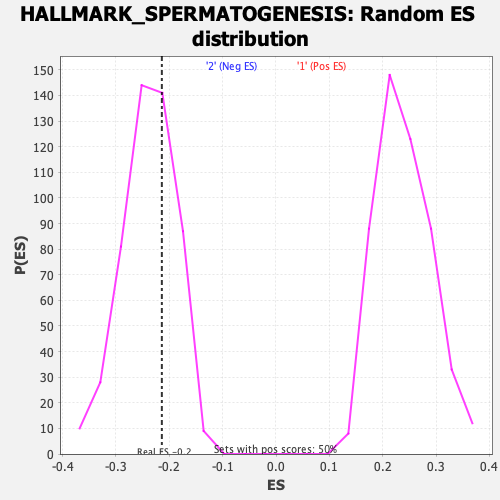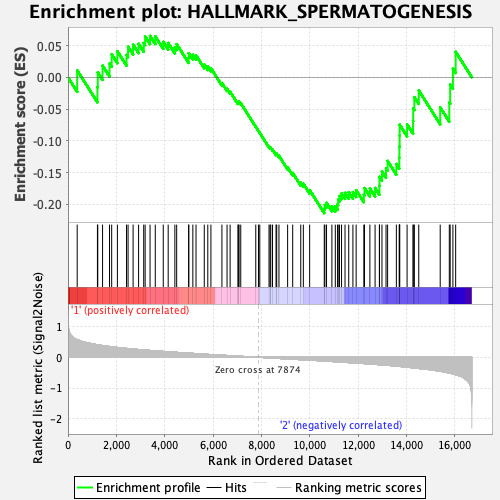
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.21389678 |
| Normalized Enrichment Score (NES) | -0.8977555 |
| Nominal p-value | 0.622 |
| FDR q-value | 0.9784894 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zc3h14 | 380 | 0.575 | 0.0110 | No |
| 2 | Pacrg | 1217 | 0.407 | -0.0153 | No |
| 3 | Ide | 1231 | 0.405 | 0.0078 | No |
| 4 | Nphp1 | 1429 | 0.381 | 0.0184 | No |
| 5 | Arl4a | 1717 | 0.349 | 0.0217 | No |
| 6 | Ldhc | 1806 | 0.339 | 0.0364 | No |
| 7 | Adam2 | 2042 | 0.319 | 0.0411 | No |
| 8 | Vdac3 | 2420 | 0.288 | 0.0354 | No |
| 9 | Tcp11 | 2484 | 0.284 | 0.0483 | No |
| 10 | Alox15 | 2692 | 0.267 | 0.0516 | No |
| 11 | Pebp1 | 2919 | 0.251 | 0.0528 | No |
| 12 | Ddx4 | 3131 | 0.237 | 0.0541 | No |
| 13 | Camk4 | 3190 | 0.234 | 0.0644 | No |
| 14 | Tnni3 | 3393 | 0.221 | 0.0653 | No |
| 15 | Gfi1 | 3609 | 0.207 | 0.0645 | No |
| 16 | Nf2 | 3939 | 0.190 | 0.0560 | No |
| 17 | Mtor | 4143 | 0.177 | 0.0542 | No |
| 18 | Sirt1 | 4419 | 0.165 | 0.0473 | No |
| 19 | Cct6b | 4494 | 0.160 | 0.0523 | No |
| 20 | Acrbp | 4989 | 0.134 | 0.0305 | No |
| 21 | Dnajb8 | 4995 | 0.134 | 0.0381 | No |
| 22 | Phkg2 | 5163 | 0.126 | 0.0355 | No |
| 23 | Pgs1 | 5294 | 0.119 | 0.0347 | No |
| 24 | Ip6k1 | 5635 | 0.101 | 0.0201 | No |
| 25 | Strbp | 5781 | 0.094 | 0.0169 | No |
| 26 | Grm8 | 5910 | 0.088 | 0.0144 | No |
| 27 | Hspa4l | 6365 | 0.068 | -0.0089 | No |
| 28 | Lpin1 | 6577 | 0.058 | -0.0182 | No |
| 29 | Tle4 | 6704 | 0.052 | -0.0227 | No |
| 30 | Rfc4 | 7026 | 0.037 | -0.0399 | No |
| 31 | Ybx2 | 7059 | 0.035 | -0.0397 | No |
| 32 | Jam3 | 7069 | 0.035 | -0.0382 | No |
| 33 | Psmg1 | 7144 | 0.032 | -0.0408 | No |
| 34 | Nefh | 7765 | 0.004 | -0.0778 | No |
| 35 | Nos1 | 7877 | 0.000 | -0.0845 | No |
| 36 | Kif2c | 7931 | -0.001 | -0.0876 | No |
| 37 | Ift88 | 8313 | -0.019 | -0.1094 | No |
| 38 | Hspa2 | 8356 | -0.021 | -0.1107 | No |
| 39 | Gstm5 | 8385 | -0.022 | -0.1111 | No |
| 40 | Tulp2 | 8458 | -0.026 | -0.1139 | No |
| 41 | Prkar2a | 8603 | -0.033 | -0.1206 | No |
| 42 | Agfg1 | 8631 | -0.034 | -0.1203 | No |
| 43 | Pias2 | 8724 | -0.038 | -0.1236 | No |
| 44 | Ccnb2 | 9082 | -0.055 | -0.1418 | No |
| 45 | Clpb | 9292 | -0.064 | -0.1506 | No |
| 46 | Il12rb2 | 9625 | -0.078 | -0.1660 | No |
| 47 | Stam2 | 9732 | -0.083 | -0.1675 | No |
| 48 | Zc2hc1c | 9993 | -0.094 | -0.1776 | No |
| 49 | Csnk2a2 | 10597 | -0.121 | -0.2068 | Yes |
| 50 | Coil | 10623 | -0.122 | -0.2011 | Yes |
| 51 | Septin4 | 10691 | -0.125 | -0.1977 | Yes |
| 52 | Ttk | 10913 | -0.136 | -0.2030 | Yes |
| 53 | Cdk1 | 11054 | -0.143 | -0.2030 | Yes |
| 54 | Taldo1 | 11148 | -0.147 | -0.1999 | Yes |
| 55 | Parp2 | 11172 | -0.148 | -0.1926 | Yes |
| 56 | Mllt10 | 11227 | -0.151 | -0.1869 | Yes |
| 57 | Dbf4 | 11312 | -0.155 | -0.1829 | Yes |
| 58 | Nek2 | 11456 | -0.163 | -0.1819 | Yes |
| 59 | Rad17 | 11609 | -0.172 | -0.1809 | Yes |
| 60 | Topbp1 | 11782 | -0.180 | -0.1806 | Yes |
| 61 | Cnih2 | 11916 | -0.186 | -0.1776 | Yes |
| 62 | Ddx25 | 12234 | -0.203 | -0.1848 | Yes |
| 63 | Clvs1 | 12261 | -0.205 | -0.1743 | Yes |
| 64 | Slc12a2 | 12484 | -0.217 | -0.1748 | Yes |
| 65 | Ncaph | 12699 | -0.229 | -0.1742 | Yes |
| 66 | Spata6 | 12877 | -0.240 | -0.1707 | Yes |
| 67 | Gmcl1 | 12881 | -0.241 | -0.1567 | Yes |
| 68 | Scg5 | 12983 | -0.247 | -0.1482 | Yes |
| 69 | Map7 | 13152 | -0.257 | -0.1432 | Yes |
| 70 | Aurka | 13217 | -0.262 | -0.1316 | Yes |
| 71 | Chfr | 13579 | -0.287 | -0.1364 | Yes |
| 72 | Mlf1 | 13700 | -0.296 | -0.1262 | Yes |
| 73 | Tsn | 13704 | -0.296 | -0.1090 | Yes |
| 74 | Cdkn3 | 13713 | -0.296 | -0.0920 | Yes |
| 75 | Ezh2 | 13716 | -0.297 | -0.0746 | Yes |
| 76 | Art3 | 14022 | -0.320 | -0.0741 | Yes |
| 77 | Braf | 14270 | -0.342 | -0.0688 | Yes |
| 78 | Zpbp | 14276 | -0.343 | -0.0488 | Yes |
| 79 | Phf7 | 14323 | -0.348 | -0.0311 | Yes |
| 80 | Dcc | 14505 | -0.362 | -0.0207 | Yes |
| 81 | Dmc1 | 15392 | -0.453 | -0.0473 | Yes |
| 82 | Pcsk4 | 15768 | -0.512 | -0.0397 | Yes |
| 83 | Mast2 | 15804 | -0.518 | -0.0113 | Yes |
| 84 | Bub1 | 15917 | -0.536 | 0.0135 | Yes |
| 85 | Gapdhs | 16031 | -0.568 | 0.0402 | Yes |