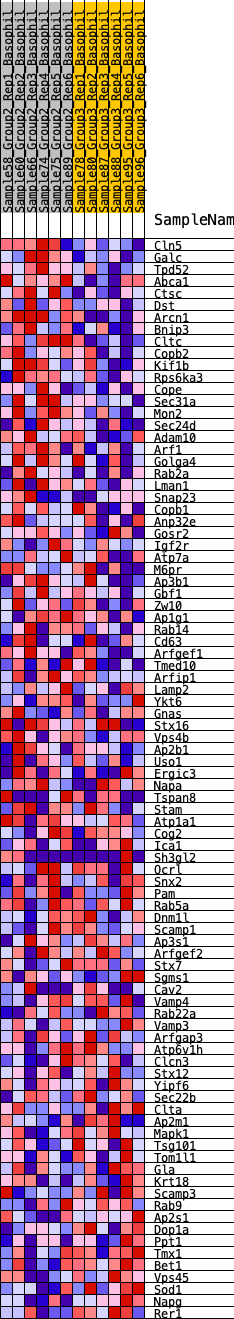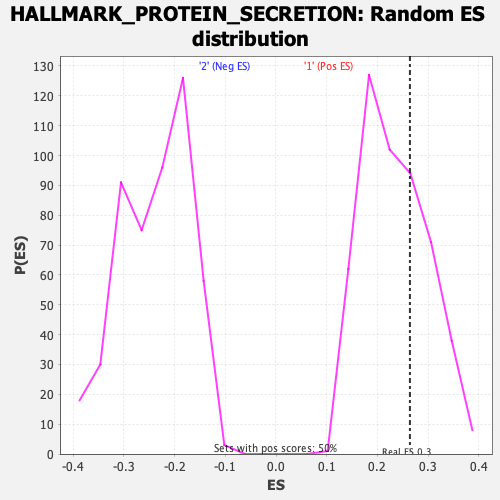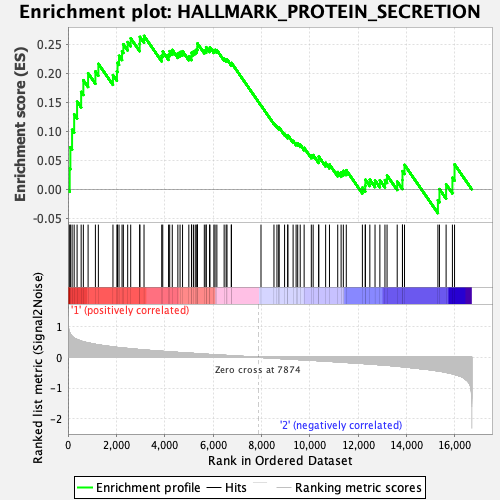
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.26421344 |
| Normalized Enrichment Score (NES) | 1.1264324 |
| Nominal p-value | 0.3280318 |
| FDR q-value | 0.5781534 |
| FWER p-Value | 0.971 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cln5 | 70 | 0.795 | 0.0361 | Yes |
| 2 | Galc | 98 | 0.750 | 0.0725 | Yes |
| 3 | Tpd52 | 169 | 0.679 | 0.1027 | Yes |
| 4 | Abca1 | 255 | 0.622 | 0.1292 | Yes |
| 5 | Ctsc | 376 | 0.576 | 0.1511 | Yes |
| 6 | Dst | 544 | 0.527 | 0.1678 | Yes |
| 7 | Arcn1 | 635 | 0.502 | 0.1879 | Yes |
| 8 | Bnip3 | 831 | 0.468 | 0.1999 | Yes |
| 9 | Cltc | 1133 | 0.419 | 0.2030 | Yes |
| 10 | Copb2 | 1256 | 0.402 | 0.2161 | Yes |
| 11 | Kif1b | 1859 | 0.333 | 0.1967 | Yes |
| 12 | Rps6ka3 | 2023 | 0.319 | 0.2031 | Yes |
| 13 | Cope | 2048 | 0.318 | 0.2178 | Yes |
| 14 | Sec31a | 2110 | 0.312 | 0.2300 | Yes |
| 15 | Mon2 | 2235 | 0.302 | 0.2379 | Yes |
| 16 | Sec24d | 2289 | 0.299 | 0.2498 | Yes |
| 17 | Adam10 | 2468 | 0.285 | 0.2536 | Yes |
| 18 | Arf1 | 2593 | 0.274 | 0.2600 | Yes |
| 19 | Golga4 | 2964 | 0.249 | 0.2503 | Yes |
| 20 | Rab2a | 2972 | 0.248 | 0.2625 | Yes |
| 21 | Lman1 | 3144 | 0.237 | 0.2642 | Yes |
| 22 | Snap23 | 3876 | 0.194 | 0.2301 | No |
| 23 | Copb1 | 3918 | 0.191 | 0.2373 | No |
| 24 | Anp32e | 4159 | 0.176 | 0.2318 | No |
| 25 | Gosr2 | 4205 | 0.174 | 0.2379 | No |
| 26 | Igf2r | 4312 | 0.169 | 0.2401 | No |
| 27 | Atp7a | 4543 | 0.157 | 0.2342 | No |
| 28 | M6pr | 4634 | 0.153 | 0.2365 | No |
| 29 | Ap3b1 | 4737 | 0.147 | 0.2378 | No |
| 30 | Gbf1 | 4997 | 0.134 | 0.2290 | No |
| 31 | Zw10 | 5106 | 0.129 | 0.2290 | No |
| 32 | Ap1g1 | 5111 | 0.129 | 0.2353 | No |
| 33 | Rab14 | 5187 | 0.125 | 0.2372 | No |
| 34 | Cd63 | 5262 | 0.121 | 0.2389 | No |
| 35 | Arfgef1 | 5324 | 0.117 | 0.2411 | No |
| 36 | Tmed10 | 5352 | 0.116 | 0.2454 | No |
| 37 | Arfip1 | 5353 | 0.115 | 0.2512 | No |
| 38 | Lamp2 | 5637 | 0.100 | 0.2393 | No |
| 39 | Ykt6 | 5711 | 0.097 | 0.2398 | No |
| 40 | Gnas | 5712 | 0.097 | 0.2447 | No |
| 41 | Stx16 | 5853 | 0.090 | 0.2409 | No |
| 42 | Vps4b | 5870 | 0.090 | 0.2445 | No |
| 43 | Ap2b1 | 6033 | 0.082 | 0.2389 | No |
| 44 | Uso1 | 6081 | 0.080 | 0.2402 | No |
| 45 | Ergic3 | 6156 | 0.077 | 0.2396 | No |
| 46 | Napa | 6457 | 0.064 | 0.2248 | No |
| 47 | Tspan8 | 6538 | 0.060 | 0.2230 | No |
| 48 | Stam | 6575 | 0.058 | 0.2238 | No |
| 49 | Atp1a1 | 6751 | 0.049 | 0.2157 | No |
| 50 | Cog2 | 6761 | 0.049 | 0.2177 | No |
| 51 | Ica1 | 7981 | -0.004 | 0.1445 | No |
| 52 | Sh3gl2 | 8517 | -0.028 | 0.1137 | No |
| 53 | Ocrl | 8641 | -0.034 | 0.1080 | No |
| 54 | Snx2 | 8719 | -0.038 | 0.1053 | No |
| 55 | Pam | 8727 | -0.038 | 0.1068 | No |
| 56 | Rab5a | 8956 | -0.049 | 0.0956 | No |
| 57 | Dnm1l | 9087 | -0.055 | 0.0905 | No |
| 58 | Scamp1 | 9090 | -0.056 | 0.0932 | No |
| 59 | Ap3s1 | 9309 | -0.064 | 0.0834 | No |
| 60 | Arfgef2 | 9433 | -0.069 | 0.0795 | No |
| 61 | Stx7 | 9499 | -0.072 | 0.0792 | No |
| 62 | Sgms1 | 9603 | -0.077 | 0.0769 | No |
| 63 | Cav2 | 9765 | -0.084 | 0.0715 | No |
| 64 | Vamp4 | 10062 | -0.097 | 0.0586 | No |
| 65 | Rab22a | 10138 | -0.099 | 0.0591 | No |
| 66 | Vamp3 | 10367 | -0.110 | 0.0510 | No |
| 67 | Arfgap3 | 10368 | -0.110 | 0.0566 | No |
| 68 | Atp6v1h | 10654 | -0.124 | 0.0457 | No |
| 69 | Clcn3 | 10813 | -0.131 | 0.0428 | No |
| 70 | Stx12 | 11154 | -0.148 | 0.0298 | No |
| 71 | Yipf6 | 11294 | -0.154 | 0.0293 | No |
| 72 | Sec22b | 11386 | -0.159 | 0.0319 | No |
| 73 | Clta | 11508 | -0.166 | 0.0330 | No |
| 74 | Ap2m1 | 12174 | -0.200 | 0.0031 | No |
| 75 | Mapk1 | 12291 | -0.206 | 0.0066 | No |
| 76 | Tsg101 | 12299 | -0.207 | 0.0167 | No |
| 77 | Tom1l1 | 12482 | -0.217 | 0.0167 | No |
| 78 | Gla | 12697 | -0.229 | 0.0154 | No |
| 79 | Krt18 | 12897 | -0.242 | 0.0157 | No |
| 80 | Scamp3 | 13107 | -0.254 | 0.0160 | No |
| 81 | Rab9 | 13194 | -0.260 | 0.0240 | No |
| 82 | Ap2s1 | 13614 | -0.289 | 0.0135 | No |
| 83 | Dop1a | 13830 | -0.304 | 0.0160 | No |
| 84 | Ppt1 | 13833 | -0.305 | 0.0313 | No |
| 85 | Tmx1 | 13916 | -0.311 | 0.0421 | No |
| 86 | Bet1 | 15297 | -0.440 | -0.0186 | No |
| 87 | Vps45 | 15361 | -0.451 | 0.0005 | No |
| 88 | Sod1 | 15636 | -0.488 | 0.0087 | No |
| 89 | Napg | 15897 | -0.533 | 0.0201 | No |
| 90 | Rer1 | 15985 | -0.555 | 0.0430 | No |