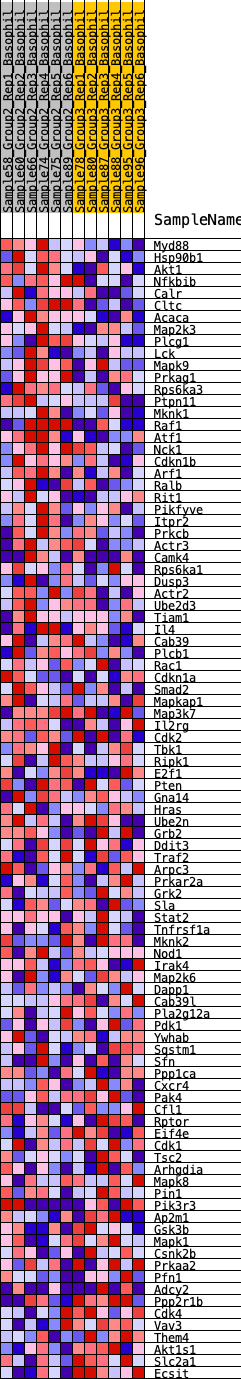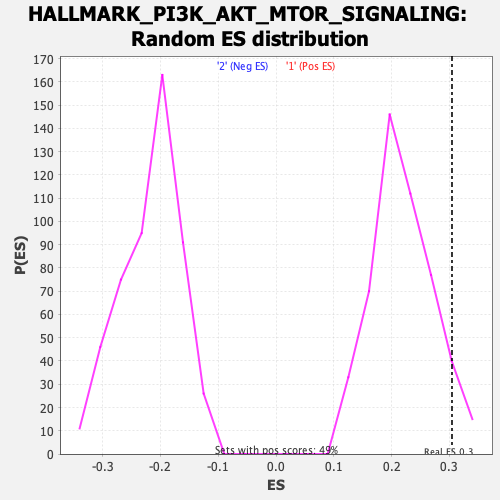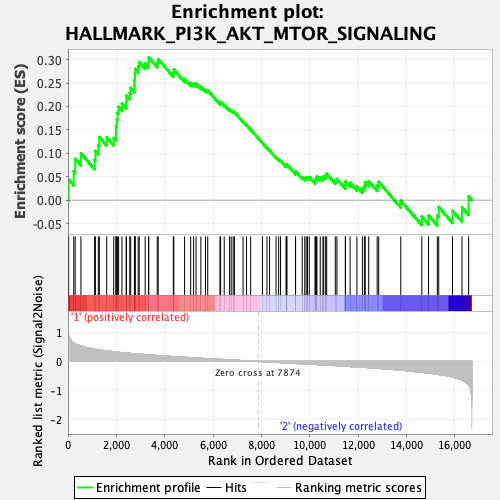
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.30467588 |
| Normalized Enrichment Score (NES) | 1.3847413 |
| Nominal p-value | 0.06896552 |
| FDR q-value | 0.45494407 |
| FWER p-Value | 0.683 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myd88 | 27 | 0.925 | 0.0437 | Yes |
| 2 | Hsp90b1 | 237 | 0.627 | 0.0618 | Yes |
| 3 | Akt1 | 297 | 0.606 | 0.0879 | Yes |
| 4 | Nfkbib | 535 | 0.530 | 0.0996 | Yes |
| 5 | Calr | 1101 | 0.424 | 0.0864 | Yes |
| 6 | Cltc | 1133 | 0.419 | 0.1050 | Yes |
| 7 | Acaca | 1254 | 0.403 | 0.1175 | Yes |
| 8 | Map2k3 | 1295 | 0.397 | 0.1346 | Yes |
| 9 | Plcg1 | 1601 | 0.363 | 0.1340 | Yes |
| 10 | Lck | 1891 | 0.331 | 0.1328 | Yes |
| 11 | Mapk9 | 1982 | 0.324 | 0.1432 | Yes |
| 12 | Prkag1 | 1987 | 0.323 | 0.1588 | Yes |
| 13 | Rps6ka3 | 2023 | 0.319 | 0.1724 | Yes |
| 14 | Ptpn11 | 2051 | 0.318 | 0.1863 | Yes |
| 15 | Mknk1 | 2093 | 0.313 | 0.1992 | Yes |
| 16 | Raf1 | 2230 | 0.303 | 0.2058 | Yes |
| 17 | Atf1 | 2406 | 0.289 | 0.2095 | Yes |
| 18 | Nck1 | 2414 | 0.289 | 0.2232 | Yes |
| 19 | Cdkn1b | 2553 | 0.278 | 0.2285 | Yes |
| 20 | Arf1 | 2593 | 0.274 | 0.2396 | Yes |
| 21 | Ralb | 2747 | 0.264 | 0.2433 | Yes |
| 22 | Rit1 | 2751 | 0.264 | 0.2560 | Yes |
| 23 | Pikfyve | 2770 | 0.262 | 0.2677 | Yes |
| 24 | Itpr2 | 2779 | 0.261 | 0.2801 | Yes |
| 25 | Prkcb | 2901 | 0.253 | 0.2851 | Yes |
| 26 | Actr3 | 2945 | 0.249 | 0.2948 | Yes |
| 27 | Camk4 | 3190 | 0.234 | 0.2915 | Yes |
| 28 | Rps6ka1 | 3332 | 0.225 | 0.2940 | Yes |
| 29 | Dusp3 | 3339 | 0.224 | 0.3047 | Yes |
| 30 | Actr2 | 3689 | 0.202 | 0.2936 | No |
| 31 | Ube2d3 | 3730 | 0.200 | 0.3010 | No |
| 32 | Tiam1 | 4355 | 0.168 | 0.2716 | No |
| 33 | Il4 | 4376 | 0.167 | 0.2786 | No |
| 34 | Cab39 | 4821 | 0.142 | 0.2588 | No |
| 35 | Plcb1 | 5073 | 0.130 | 0.2501 | No |
| 36 | Rac1 | 5194 | 0.125 | 0.2490 | No |
| 37 | Cdkn1a | 5296 | 0.119 | 0.2487 | No |
| 38 | Smad2 | 5494 | 0.108 | 0.2421 | No |
| 39 | Mapkap1 | 5686 | 0.098 | 0.2354 | No |
| 40 | Map3k7 | 5778 | 0.094 | 0.2346 | No |
| 41 | Il2rg | 6283 | 0.072 | 0.2077 | No |
| 42 | Cdk2 | 6313 | 0.071 | 0.2095 | No |
| 43 | Tbk1 | 6460 | 0.063 | 0.2038 | No |
| 44 | Ripk1 | 6682 | 0.053 | 0.1930 | No |
| 45 | E2f1 | 6747 | 0.050 | 0.1916 | No |
| 46 | Pten | 6829 | 0.046 | 0.1890 | No |
| 47 | Gna14 | 6882 | 0.044 | 0.1880 | No |
| 48 | Hras | 7241 | 0.027 | 0.1678 | No |
| 49 | Ube2n | 7381 | 0.021 | 0.1604 | No |
| 50 | Grb2 | 7554 | 0.014 | 0.1508 | No |
| 51 | Ddit3 | 8041 | -0.006 | 0.1218 | No |
| 52 | Traf2 | 8228 | -0.015 | 0.1114 | No |
| 53 | Arpc3 | 8336 | -0.020 | 0.1059 | No |
| 54 | Prkar2a | 8603 | -0.033 | 0.0915 | No |
| 55 | Grk2 | 8711 | -0.037 | 0.0869 | No |
| 56 | Sla | 8788 | -0.041 | 0.0843 | No |
| 57 | Stat2 | 9017 | -0.052 | 0.0731 | No |
| 58 | Tnfrsf1a | 9035 | -0.053 | 0.0747 | No |
| 59 | Mknk2 | 9055 | -0.054 | 0.0762 | No |
| 60 | Nod1 | 9409 | -0.068 | 0.0582 | No |
| 61 | Irak4 | 9411 | -0.068 | 0.0615 | No |
| 62 | Map2k6 | 9685 | -0.081 | 0.0490 | No |
| 63 | Dapp1 | 9791 | -0.085 | 0.0468 | No |
| 64 | Cab39l | 9848 | -0.087 | 0.0477 | No |
| 65 | Pla2g12a | 9904 | -0.090 | 0.0488 | No |
| 66 | Pdk1 | 9978 | -0.093 | 0.0490 | No |
| 67 | Ywhab | 10215 | -0.103 | 0.0398 | No |
| 68 | Sqstm1 | 10238 | -0.104 | 0.0436 | No |
| 69 | Sfn | 10268 | -0.105 | 0.0470 | No |
| 70 | Ppp1ca | 10294 | -0.106 | 0.0507 | No |
| 71 | Cxcr4 | 10431 | -0.113 | 0.0480 | No |
| 72 | Pak4 | 10542 | -0.119 | 0.0472 | No |
| 73 | Cfl1 | 10575 | -0.120 | 0.0512 | No |
| 74 | Rptor | 10657 | -0.124 | 0.0524 | No |
| 75 | Eif4e | 10697 | -0.126 | 0.0562 | No |
| 76 | Cdk1 | 11054 | -0.143 | 0.0417 | No |
| 77 | Tsc2 | 11120 | -0.146 | 0.0450 | No |
| 78 | Arhgdia | 11469 | -0.163 | 0.0320 | No |
| 79 | Mapk8 | 11475 | -0.164 | 0.0397 | No |
| 80 | Pin1 | 11667 | -0.175 | 0.0368 | No |
| 81 | Pik3r3 | 11948 | -0.188 | 0.0291 | No |
| 82 | Ap2m1 | 12174 | -0.200 | 0.0254 | No |
| 83 | Gsk3b | 12257 | -0.205 | 0.0305 | No |
| 84 | Mapk1 | 12291 | -0.206 | 0.0386 | No |
| 85 | Csnk2b | 12435 | -0.214 | 0.0405 | No |
| 86 | Prkaa2 | 12785 | -0.234 | 0.0309 | No |
| 87 | Pfn1 | 12849 | -0.239 | 0.0388 | No |
| 88 | Adcy2 | 13762 | -0.300 | -0.0014 | No |
| 89 | Ppp2r1b | 14632 | -0.374 | -0.0354 | No |
| 90 | Cdk4 | 14909 | -0.397 | -0.0326 | No |
| 91 | Vav3 | 15273 | -0.438 | -0.0330 | No |
| 92 | Them4 | 15331 | -0.445 | -0.0147 | No |
| 93 | Akt1s1 | 15901 | -0.533 | -0.0229 | No |
| 94 | Slc2a1 | 16294 | -0.638 | -0.0152 | No |
| 95 | Ecsit | 16568 | -0.807 | 0.0079 | No |