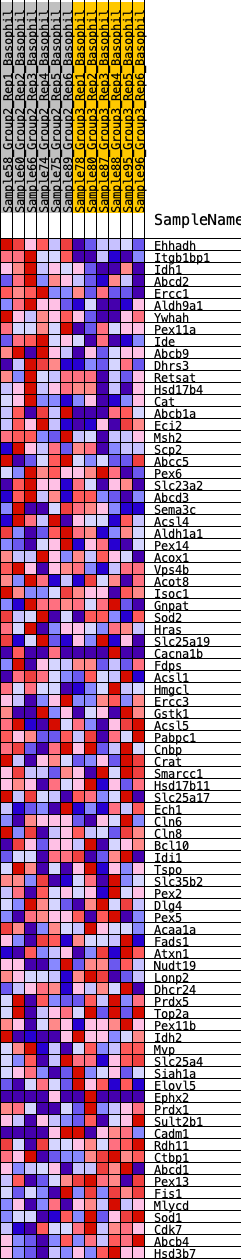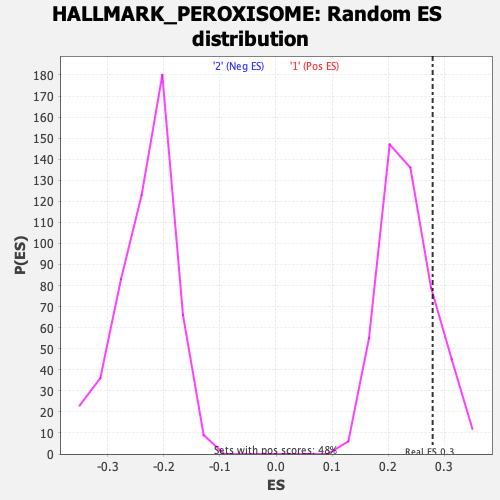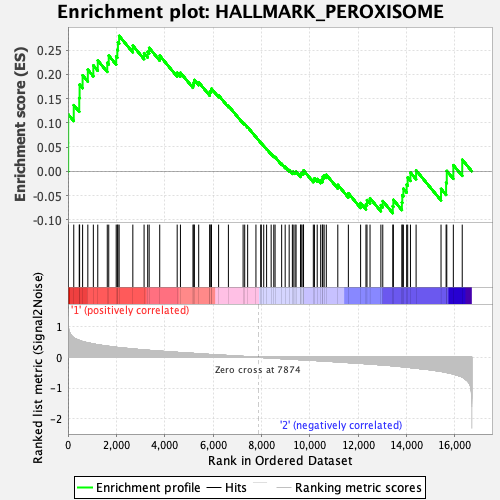
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.27914914 |
| Normalized Enrichment Score (NES) | 1.189248 |
| Nominal p-value | 0.1875 |
| FDR q-value | 0.7003543 |
| FWER p-Value | 0.94 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ehhadh | 7 | 1.132 | 0.0586 | Yes |
| 2 | Itgb1bp1 | 8 | 1.127 | 0.1174 | Yes |
| 3 | Idh1 | 238 | 0.627 | 0.1363 | Yes |
| 4 | Abcd2 | 465 | 0.547 | 0.1512 | Yes |
| 5 | Ercc1 | 480 | 0.543 | 0.1787 | Yes |
| 6 | Aldh9a1 | 604 | 0.510 | 0.1978 | Yes |
| 7 | Ywhah | 820 | 0.470 | 0.2094 | Yes |
| 8 | Pex11a | 1048 | 0.432 | 0.2183 | Yes |
| 9 | Ide | 1231 | 0.405 | 0.2284 | Yes |
| 10 | Abcb9 | 1624 | 0.361 | 0.2237 | Yes |
| 11 | Dhrs3 | 1685 | 0.353 | 0.2385 | Yes |
| 12 | Retsat | 1993 | 0.323 | 0.2368 | Yes |
| 13 | Hsd17b4 | 2040 | 0.319 | 0.2507 | Yes |
| 14 | Cat | 2065 | 0.317 | 0.2658 | Yes |
| 15 | Abcb1a | 2114 | 0.312 | 0.2791 | Yes |
| 16 | Eci2 | 2681 | 0.268 | 0.2591 | No |
| 17 | Msh2 | 3146 | 0.237 | 0.2435 | No |
| 18 | Scp2 | 3296 | 0.226 | 0.2463 | No |
| 19 | Abcc5 | 3356 | 0.224 | 0.2544 | No |
| 20 | Pex6 | 3793 | 0.198 | 0.2385 | No |
| 21 | Slc23a2 | 4515 | 0.159 | 0.2033 | No |
| 22 | Abcd3 | 4650 | 0.152 | 0.2032 | No |
| 23 | Sema3c | 5168 | 0.126 | 0.1787 | No |
| 24 | Acsl4 | 5202 | 0.124 | 0.1832 | No |
| 25 | Aldh1a1 | 5225 | 0.123 | 0.1883 | No |
| 26 | Pex14 | 5405 | 0.113 | 0.1834 | No |
| 27 | Acox1 | 5863 | 0.090 | 0.1606 | No |
| 28 | Vps4b | 5870 | 0.090 | 0.1649 | No |
| 29 | Acot8 | 5918 | 0.087 | 0.1666 | No |
| 30 | Isoc1 | 5933 | 0.087 | 0.1703 | No |
| 31 | Gnpat | 6228 | 0.074 | 0.1565 | No |
| 32 | Sod2 | 6633 | 0.055 | 0.1350 | No |
| 33 | Hras | 7241 | 0.027 | 0.0999 | No |
| 34 | Slc25a19 | 7307 | 0.025 | 0.0973 | No |
| 35 | Cacna1b | 7429 | 0.019 | 0.0910 | No |
| 36 | Fdps | 7772 | 0.004 | 0.0706 | No |
| 37 | Acsl1 | 7971 | -0.003 | 0.0589 | No |
| 38 | Hmgcl | 7992 | -0.004 | 0.0579 | No |
| 39 | Ercc3 | 8094 | -0.009 | 0.0523 | No |
| 40 | Gstk1 | 8213 | -0.015 | 0.0460 | No |
| 41 | Acsl5 | 8401 | -0.023 | 0.0359 | No |
| 42 | Pabpc1 | 8510 | -0.028 | 0.0309 | No |
| 43 | Cnbp | 8563 | -0.030 | 0.0294 | No |
| 44 | Crat | 8835 | -0.044 | 0.0153 | No |
| 45 | Smarcc1 | 8981 | -0.050 | 0.0092 | No |
| 46 | Hsd17b11 | 9146 | -0.058 | 0.0024 | No |
| 47 | Slc25a17 | 9278 | -0.063 | -0.0022 | No |
| 48 | Ech1 | 9310 | -0.064 | -0.0007 | No |
| 49 | Cln6 | 9387 | -0.067 | -0.0018 | No |
| 50 | Cln8 | 9431 | -0.069 | -0.0008 | No |
| 51 | Bcl10 | 9614 | -0.078 | -0.0077 | No |
| 52 | Idi1 | 9634 | -0.078 | -0.0048 | No |
| 53 | Tspo | 9676 | -0.080 | -0.0030 | No |
| 54 | Slc35b2 | 9733 | -0.083 | -0.0021 | No |
| 55 | Pex2 | 9740 | -0.083 | 0.0019 | No |
| 56 | Dlg4 | 10145 | -0.100 | -0.0173 | No |
| 57 | Pex5 | 10191 | -0.102 | -0.0147 | No |
| 58 | Acaa1a | 10306 | -0.107 | -0.0159 | No |
| 59 | Fads1 | 10449 | -0.114 | -0.0185 | No |
| 60 | Atxn1 | 10518 | -0.118 | -0.0165 | No |
| 61 | Nudt19 | 10537 | -0.119 | -0.0114 | No |
| 62 | Lonp2 | 10592 | -0.121 | -0.0083 | No |
| 63 | Dhcr24 | 10682 | -0.125 | -0.0072 | No |
| 64 | Prdx5 | 11157 | -0.148 | -0.0280 | No |
| 65 | Top2a | 11594 | -0.171 | -0.0453 | No |
| 66 | Pex11b | 12100 | -0.196 | -0.0655 | No |
| 67 | Idh2 | 12322 | -0.208 | -0.0680 | No |
| 68 | Mvp | 12364 | -0.211 | -0.0595 | No |
| 69 | Slc25a4 | 12490 | -0.217 | -0.0557 | No |
| 70 | Siah1a | 12939 | -0.245 | -0.0698 | No |
| 71 | Elovl5 | 13017 | -0.248 | -0.0615 | No |
| 72 | Ephx2 | 13431 | -0.276 | -0.0720 | No |
| 73 | Prdx1 | 13455 | -0.278 | -0.0589 | No |
| 74 | Sult2b1 | 13808 | -0.303 | -0.0642 | No |
| 75 | Cadm1 | 13824 | -0.304 | -0.0493 | No |
| 76 | Rdh11 | 13869 | -0.308 | -0.0359 | No |
| 77 | Ctbp1 | 14005 | -0.319 | -0.0274 | No |
| 78 | Abcd1 | 14047 | -0.322 | -0.0131 | No |
| 79 | Pex13 | 14162 | -0.333 | -0.0025 | No |
| 80 | Fis1 | 14395 | -0.353 | 0.0019 | No |
| 81 | Mlycd | 15426 | -0.457 | -0.0363 | No |
| 82 | Sod1 | 15636 | -0.488 | -0.0234 | No |
| 83 | Cdk7 | 15664 | -0.493 | 0.0007 | No |
| 84 | Abcb4 | 15935 | -0.539 | 0.0125 | No |
| 85 | Hsd3b7 | 16303 | -0.640 | 0.0238 | No |