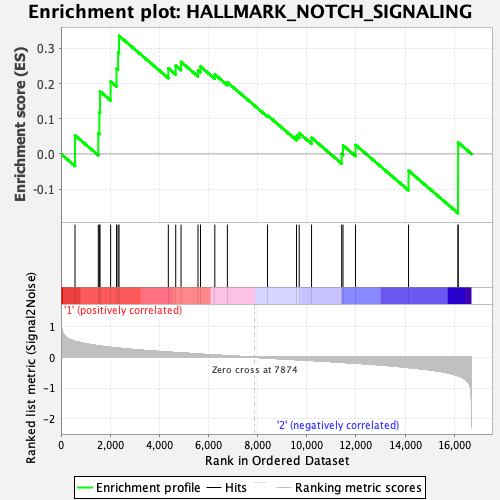
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | 0.3357375 |
| Normalized Enrichment Score (NES) | 0.93282753 |
| Nominal p-value | 0.53137255 |
| FDR q-value | 0.8820348 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fzd7 | 570 | 0.516 | 0.0530 | Yes |
| 2 | Psen2 | 1517 | 0.371 | 0.0588 | Yes |
| 3 | Jag1 | 1560 | 0.366 | 0.1181 | Yes |
| 4 | Prkca | 1588 | 0.364 | 0.1779 | Yes |
| 5 | Ppard | 2019 | 0.320 | 0.2060 | Yes |
| 6 | Lfng | 2259 | 0.301 | 0.2425 | Yes |
| 7 | Dtx4 | 2320 | 0.296 | 0.2888 | Yes |
| 8 | Cul1 | 2361 | 0.292 | 0.3357 | Yes |
| 9 | Notch3 | 4366 | 0.167 | 0.2438 | No |
| 10 | Tcf7l2 | 4667 | 0.151 | 0.2512 | No |
| 11 | Notch2 | 4883 | 0.139 | 0.2618 | No |
| 12 | St3gal6 | 5574 | 0.104 | 0.2379 | No |
| 13 | Sap30 | 5675 | 0.099 | 0.2486 | No |
| 14 | Fbxw11 | 6258 | 0.073 | 0.2261 | No |
| 15 | Notch1 | 6768 | 0.048 | 0.2037 | No |
| 16 | Dtx2 | 8402 | -0.023 | 0.1098 | No |
| 17 | Arrb1 | 9582 | -0.076 | 0.0518 | No |
| 18 | Rbx1 | 9691 | -0.081 | 0.0590 | No |
| 19 | Psenen | 10193 | -0.102 | 0.0462 | No |
| 20 | Skp1 | 11418 | -0.161 | -0.0001 | No |
| 21 | Aph1a | 11473 | -0.164 | 0.0243 | No |
| 22 | Fzd5 | 11982 | -0.190 | 0.0259 | No |
| 23 | Kat2a | 14139 | -0.331 | -0.0476 | No |
| 24 | Maml2 | 16151 | -0.595 | -0.0677 | No |
| 25 | Ccnd1 | 16155 | -0.596 | 0.0326 | No |