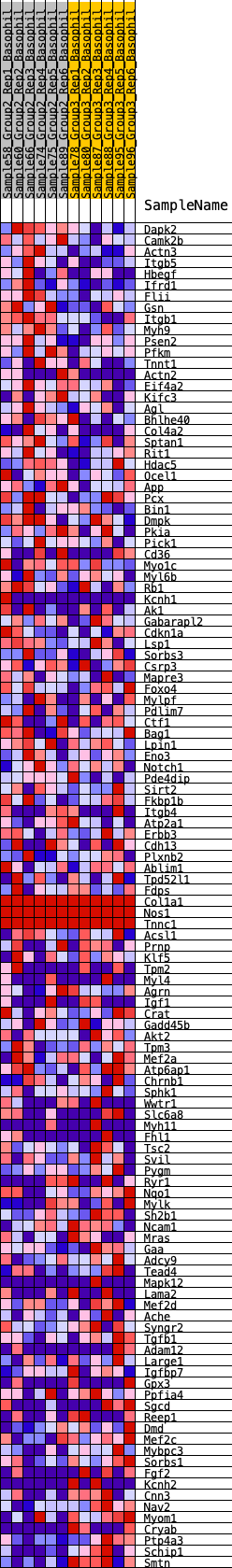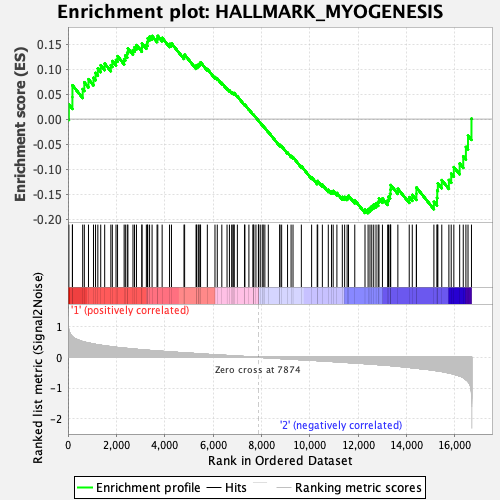
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.1881046 |
| Normalized Enrichment Score (NES) | -0.7570237 |
| Nominal p-value | 0.94860816 |
| FDR q-value | 0.98851496 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dapk2 | 36 | 0.880 | 0.0289 | No |
| 2 | Camk2b | 182 | 0.671 | 0.0439 | No |
| 3 | Actn3 | 184 | 0.669 | 0.0675 | No |
| 4 | Itgb5 | 608 | 0.508 | 0.0600 | No |
| 5 | Hbegf | 675 | 0.495 | 0.0735 | No |
| 6 | Ifrd1 | 847 | 0.466 | 0.0797 | No |
| 7 | Flii | 1054 | 0.431 | 0.0825 | No |
| 8 | Gsn | 1139 | 0.419 | 0.0922 | No |
| 9 | Itgb1 | 1234 | 0.405 | 0.1009 | No |
| 10 | Myh9 | 1353 | 0.392 | 0.1076 | No |
| 11 | Psen2 | 1517 | 0.371 | 0.1109 | No |
| 12 | Pfkm | 1772 | 0.343 | 0.1077 | No |
| 13 | Tnnt1 | 1833 | 0.336 | 0.1160 | No |
| 14 | Actn2 | 1986 | 0.324 | 0.1182 | No |
| 15 | Eif4a2 | 2049 | 0.318 | 0.1258 | No |
| 16 | Kifc3 | 2321 | 0.296 | 0.1199 | No |
| 17 | Agl | 2371 | 0.292 | 0.1272 | No |
| 18 | Bhlhe40 | 2449 | 0.287 | 0.1327 | No |
| 19 | Col4a2 | 2477 | 0.285 | 0.1412 | No |
| 20 | Sptan1 | 2687 | 0.267 | 0.1380 | No |
| 21 | Rit1 | 2751 | 0.264 | 0.1435 | No |
| 22 | Hdac5 | 2831 | 0.258 | 0.1479 | No |
| 23 | Ocel1 | 3054 | 0.242 | 0.1431 | No |
| 24 | App | 3059 | 0.241 | 0.1514 | No |
| 25 | Pcx | 3240 | 0.230 | 0.1486 | No |
| 26 | Bin1 | 3286 | 0.227 | 0.1540 | No |
| 27 | Dmpk | 3297 | 0.226 | 0.1614 | No |
| 28 | Pkia | 3373 | 0.222 | 0.1647 | No |
| 29 | Pick1 | 3477 | 0.216 | 0.1661 | No |
| 30 | Cd36 | 3683 | 0.203 | 0.1609 | No |
| 31 | Myo1c | 3713 | 0.201 | 0.1663 | No |
| 32 | Myl6b | 3893 | 0.193 | 0.1623 | No |
| 33 | Rb1 | 4199 | 0.174 | 0.1501 | No |
| 34 | Kcnh1 | 4279 | 0.169 | 0.1513 | No |
| 35 | Ak1 | 4797 | 0.144 | 0.1252 | No |
| 36 | Gabarapl2 | 4823 | 0.142 | 0.1288 | No |
| 37 | Cdkn1a | 5296 | 0.119 | 0.1045 | No |
| 38 | Lsp1 | 5314 | 0.118 | 0.1076 | No |
| 39 | Sorbs3 | 5383 | 0.114 | 0.1075 | No |
| 40 | Csrp3 | 5410 | 0.113 | 0.1100 | No |
| 41 | Mapre3 | 5458 | 0.110 | 0.1110 | No |
| 42 | Foxo4 | 5482 | 0.109 | 0.1135 | No |
| 43 | Mylpf | 5762 | 0.095 | 0.1000 | No |
| 44 | Pdlim7 | 6078 | 0.080 | 0.0839 | No |
| 45 | Ctf1 | 6173 | 0.077 | 0.0809 | No |
| 46 | Bag1 | 6361 | 0.068 | 0.0720 | No |
| 47 | Lpin1 | 6577 | 0.058 | 0.0611 | No |
| 48 | Eno3 | 6676 | 0.053 | 0.0571 | No |
| 49 | Notch1 | 6768 | 0.048 | 0.0533 | No |
| 50 | Pde4dip | 6817 | 0.046 | 0.0520 | No |
| 51 | Sirt2 | 6859 | 0.044 | 0.0511 | No |
| 52 | Fkbp1b | 6871 | 0.044 | 0.0520 | No |
| 53 | Itgb4 | 7004 | 0.038 | 0.0454 | No |
| 54 | Atp2a1 | 7298 | 0.025 | 0.0286 | No |
| 55 | Erbb3 | 7316 | 0.024 | 0.0285 | No |
| 56 | Cdh13 | 7481 | 0.017 | 0.0192 | No |
| 57 | Plxnb2 | 7653 | 0.010 | 0.0092 | No |
| 58 | Ablim1 | 7655 | 0.010 | 0.0095 | No |
| 59 | Tpd52l1 | 7687 | 0.008 | 0.0079 | No |
| 60 | Fdps | 7772 | 0.004 | 0.0030 | No |
| 61 | Col1a1 | 7874 | 0.000 | -0.0031 | No |
| 62 | Nos1 | 7877 | 0.000 | -0.0033 | No |
| 63 | Tnnc1 | 7893 | 0.000 | -0.0042 | No |
| 64 | Acsl1 | 7971 | -0.003 | -0.0087 | No |
| 65 | Prnp | 8061 | -0.007 | -0.0138 | No |
| 66 | Klf5 | 8065 | -0.007 | -0.0137 | No |
| 67 | Tpm2 | 8141 | -0.011 | -0.0178 | No |
| 68 | Myl4 | 8284 | -0.018 | -0.0258 | No |
| 69 | Agrn | 8757 | -0.039 | -0.0529 | No |
| 70 | Igf1 | 8762 | -0.039 | -0.0517 | No |
| 71 | Crat | 8835 | -0.044 | -0.0545 | No |
| 72 | Gadd45b | 9079 | -0.055 | -0.0672 | No |
| 73 | Akt2 | 9225 | -0.062 | -0.0738 | No |
| 74 | Tpm3 | 9305 | -0.064 | -0.0763 | No |
| 75 | Mef2a | 9650 | -0.080 | -0.0942 | No |
| 76 | Atp6ap1 | 10074 | -0.097 | -0.1163 | No |
| 77 | Chrnb1 | 10305 | -0.107 | -0.1264 | No |
| 78 | Sphk1 | 10326 | -0.108 | -0.1238 | No |
| 79 | Wwtr1 | 10519 | -0.118 | -0.1312 | No |
| 80 | Slc6a8 | 10764 | -0.130 | -0.1413 | No |
| 81 | Myh11 | 10896 | -0.135 | -0.1444 | No |
| 82 | Fhl1 | 10966 | -0.139 | -0.1437 | No |
| 83 | Tsc2 | 11120 | -0.146 | -0.1478 | No |
| 84 | Svil | 11344 | -0.157 | -0.1557 | No |
| 85 | Pygm | 11440 | -0.162 | -0.1557 | No |
| 86 | Ryr1 | 11543 | -0.168 | -0.1559 | No |
| 87 | Nqo1 | 11600 | -0.171 | -0.1532 | No |
| 88 | Mylk | 11859 | -0.184 | -0.1623 | No |
| 89 | Sh2b1 | 12282 | -0.206 | -0.1804 | No |
| 90 | Ncam1 | 12410 | -0.213 | -0.1806 | Yes |
| 91 | Mras | 12488 | -0.217 | -0.1775 | Yes |
| 92 | Gaa | 12562 | -0.220 | -0.1741 | Yes |
| 93 | Adcy9 | 12643 | -0.225 | -0.1710 | Yes |
| 94 | Tead4 | 12738 | -0.231 | -0.1685 | Yes |
| 95 | Mapk12 | 12826 | -0.238 | -0.1653 | Yes |
| 96 | Lama2 | 12860 | -0.240 | -0.1589 | Yes |
| 97 | Mef2d | 13003 | -0.248 | -0.1587 | Yes |
| 98 | Ache | 13220 | -0.262 | -0.1624 | Yes |
| 99 | Syngr2 | 13254 | -0.264 | -0.1551 | Yes |
| 100 | Tgfb1 | 13319 | -0.267 | -0.1495 | Yes |
| 101 | Adam12 | 13337 | -0.269 | -0.1410 | Yes |
| 102 | Large1 | 13345 | -0.269 | -0.1319 | Yes |
| 103 | Igfbp7 | 13641 | -0.291 | -0.1394 | Yes |
| 104 | Gpx3 | 14111 | -0.328 | -0.1561 | Yes |
| 105 | Ppfia4 | 14237 | -0.340 | -0.1516 | Yes |
| 106 | Sgcd | 14406 | -0.354 | -0.1492 | Yes |
| 107 | Reep1 | 14409 | -0.354 | -0.1368 | Yes |
| 108 | Dmd | 15131 | -0.422 | -0.1654 | Yes |
| 109 | Mef2c | 15259 | -0.437 | -0.1576 | Yes |
| 110 | Mybpc3 | 15270 | -0.437 | -0.1428 | Yes |
| 111 | Sorbs1 | 15295 | -0.440 | -0.1286 | Yes |
| 112 | Fgf2 | 15457 | -0.461 | -0.1220 | Yes |
| 113 | Kcnh2 | 15751 | -0.508 | -0.1217 | Yes |
| 114 | Cnn3 | 15847 | -0.525 | -0.1089 | Yes |
| 115 | Nav2 | 15956 | -0.548 | -0.0960 | Yes |
| 116 | Myom1 | 16197 | -0.609 | -0.0890 | Yes |
| 117 | Cryab | 16345 | -0.657 | -0.0746 | Yes |
| 118 | Ptp4a3 | 16455 | -0.720 | -0.0557 | Yes |
| 119 | Schip1 | 16542 | -0.786 | -0.0331 | Yes |
| 120 | Smtn | 16685 | -1.201 | 0.0008 | Yes |