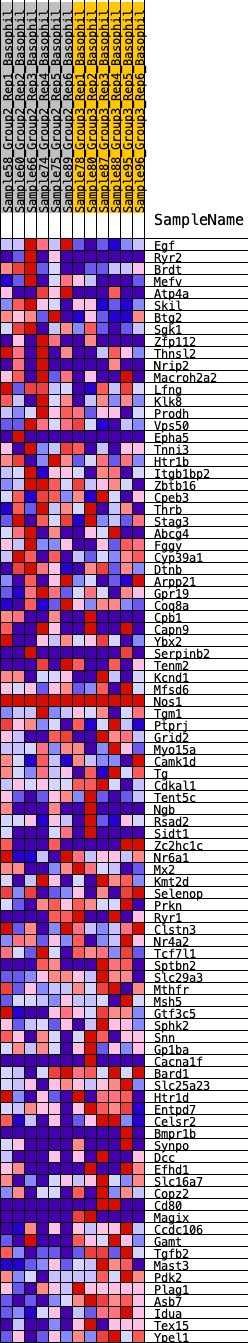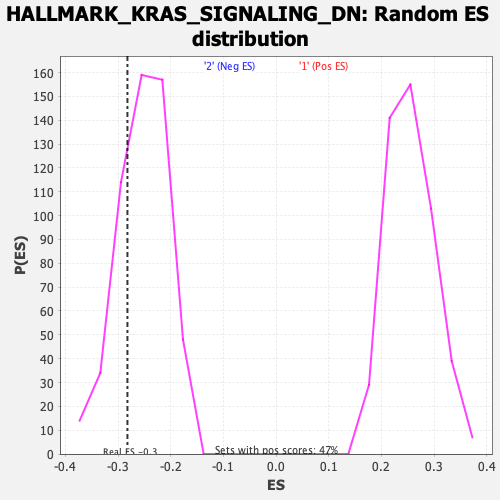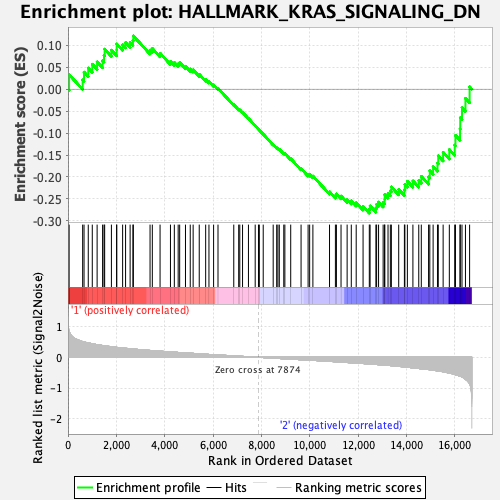
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.28233916 |
| Normalized Enrichment Score (NES) | -1.1187483 |
| Nominal p-value | 0.24144487 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.966 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Egf | 46 | 0.850 | 0.0333 | No |
| 2 | Ryr2 | 606 | 0.509 | 0.0212 | No |
| 3 | Brdt | 663 | 0.498 | 0.0390 | No |
| 4 | Mefv | 838 | 0.467 | 0.0483 | No |
| 5 | Atp4a | 1004 | 0.437 | 0.0569 | No |
| 6 | Skil | 1204 | 0.409 | 0.0623 | No |
| 7 | Btg2 | 1430 | 0.381 | 0.0649 | No |
| 8 | Sgk1 | 1489 | 0.373 | 0.0772 | No |
| 9 | Zfp112 | 1516 | 0.371 | 0.0914 | No |
| 10 | Thnsl2 | 1795 | 0.340 | 0.0890 | No |
| 11 | Nrip2 | 2012 | 0.320 | 0.0896 | No |
| 12 | Macroh2a2 | 2014 | 0.320 | 0.1031 | No |
| 13 | Lfng | 2259 | 0.301 | 0.1012 | No |
| 14 | Klk8 | 2381 | 0.291 | 0.1062 | No |
| 15 | Prodh | 2569 | 0.277 | 0.1067 | No |
| 16 | Vps50 | 2685 | 0.268 | 0.1111 | No |
| 17 | Epha5 | 2702 | 0.267 | 0.1215 | No |
| 18 | Tnni3 | 3393 | 0.221 | 0.0893 | No |
| 19 | Htr1b | 3487 | 0.215 | 0.0928 | No |
| 20 | Itgb1bp2 | 3807 | 0.197 | 0.0820 | No |
| 21 | Zbtb16 | 4237 | 0.171 | 0.0634 | No |
| 22 | Cpeb3 | 4397 | 0.166 | 0.0608 | No |
| 23 | Thrb | 4556 | 0.156 | 0.0579 | No |
| 24 | Stag3 | 4618 | 0.153 | 0.0608 | No |
| 25 | Abcg4 | 4858 | 0.141 | 0.0523 | No |
| 26 | Fggy | 5055 | 0.131 | 0.0461 | No |
| 27 | Cyp39a1 | 5175 | 0.126 | 0.0443 | No |
| 28 | Dtnb | 5427 | 0.112 | 0.0339 | No |
| 29 | Arpp21 | 5693 | 0.098 | 0.0221 | No |
| 30 | Gpr19 | 5829 | 0.092 | 0.0178 | No |
| 31 | Coq8a | 6021 | 0.083 | 0.0098 | No |
| 32 | Cpb1 | 6203 | 0.076 | 0.0022 | No |
| 33 | Capn9 | 6855 | 0.045 | -0.0351 | No |
| 34 | Ybx2 | 7059 | 0.035 | -0.0459 | No |
| 35 | Serpinb2 | 7104 | 0.034 | -0.0471 | No |
| 36 | Tenm2 | 7218 | 0.028 | -0.0527 | No |
| 37 | Kcnd1 | 7462 | 0.018 | -0.0666 | No |
| 38 | Mfsd6 | 7741 | 0.006 | -0.0831 | No |
| 39 | Nos1 | 7877 | 0.000 | -0.0912 | No |
| 40 | Tgm1 | 7912 | -0.001 | -0.0932 | No |
| 41 | Ptprj | 8073 | -0.008 | -0.1025 | No |
| 42 | Grid2 | 8485 | -0.027 | -0.1261 | No |
| 43 | Myo15a | 8630 | -0.034 | -0.1334 | No |
| 44 | Camk1d | 8680 | -0.036 | -0.1348 | No |
| 45 | Tg | 8738 | -0.038 | -0.1366 | No |
| 46 | Cdkal1 | 8923 | -0.047 | -0.1457 | No |
| 47 | Tent5c | 8968 | -0.049 | -0.1462 | No |
| 48 | Ngb | 9210 | -0.061 | -0.1582 | No |
| 49 | Rsad2 | 9638 | -0.079 | -0.1805 | No |
| 50 | Sidt1 | 9929 | -0.091 | -0.1942 | No |
| 51 | Zc2hc1c | 9993 | -0.094 | -0.1940 | No |
| 52 | Nr6a1 | 10127 | -0.099 | -0.1978 | No |
| 53 | Mx2 | 10815 | -0.131 | -0.2336 | No |
| 54 | Kmt2d | 11058 | -0.143 | -0.2421 | No |
| 55 | Selenop | 11100 | -0.145 | -0.2384 | No |
| 56 | Prkn | 11291 | -0.154 | -0.2433 | No |
| 57 | Ryr1 | 11543 | -0.168 | -0.2513 | No |
| 58 | Clstn3 | 11715 | -0.177 | -0.2541 | No |
| 59 | Nr4a2 | 11918 | -0.187 | -0.2584 | No |
| 60 | Tcf7l1 | 12202 | -0.201 | -0.2669 | No |
| 61 | Sptbn2 | 12460 | -0.215 | -0.2732 | Yes |
| 62 | Slc29a3 | 12492 | -0.217 | -0.2659 | Yes |
| 63 | Mthfr | 12733 | -0.231 | -0.2705 | Yes |
| 64 | Msh5 | 12762 | -0.233 | -0.2624 | Yes |
| 65 | Gtf3c5 | 12843 | -0.238 | -0.2571 | Yes |
| 66 | Sphk2 | 13032 | -0.250 | -0.2578 | Yes |
| 67 | Snn | 13095 | -0.253 | -0.2508 | Yes |
| 68 | Gp1ba | 13097 | -0.253 | -0.2401 | Yes |
| 69 | Cacna1f | 13234 | -0.263 | -0.2371 | Yes |
| 70 | Bard1 | 13330 | -0.268 | -0.2315 | Yes |
| 71 | Slc25a23 | 13372 | -0.271 | -0.2225 | Yes |
| 72 | Htr1d | 13676 | -0.293 | -0.2283 | Yes |
| 73 | Entpd7 | 13911 | -0.310 | -0.2292 | Yes |
| 74 | Celsr2 | 13931 | -0.312 | -0.2171 | Yes |
| 75 | Bmpr1b | 14034 | -0.321 | -0.2096 | Yes |
| 76 | Synpo | 14263 | -0.342 | -0.2089 | Yes |
| 77 | Dcc | 14505 | -0.362 | -0.2080 | Yes |
| 78 | Efhd1 | 14610 | -0.372 | -0.1985 | Yes |
| 79 | Slc16a7 | 14911 | -0.397 | -0.1997 | Yes |
| 80 | Copz2 | 14962 | -0.402 | -0.1857 | Yes |
| 81 | Cd80 | 15099 | -0.418 | -0.1762 | Yes |
| 82 | Magix | 15279 | -0.438 | -0.1684 | Yes |
| 83 | Ccdc106 | 15312 | -0.443 | -0.1516 | Yes |
| 84 | Gamt | 15513 | -0.467 | -0.1438 | Yes |
| 85 | Tgfb2 | 15769 | -0.512 | -0.1374 | Yes |
| 86 | Mast3 | 15992 | -0.556 | -0.1272 | Yes |
| 87 | Pdk2 | 16023 | -0.565 | -0.1051 | Yes |
| 88 | Plag1 | 16211 | -0.611 | -0.0904 | Yes |
| 89 | Asb7 | 16221 | -0.614 | -0.0649 | Yes |
| 90 | Idua | 16293 | -0.638 | -0.0421 | Yes |
| 91 | Tex15 | 16436 | -0.708 | -0.0207 | Yes |
| 92 | Ypel1 | 16608 | -0.859 | 0.0055 | Yes |