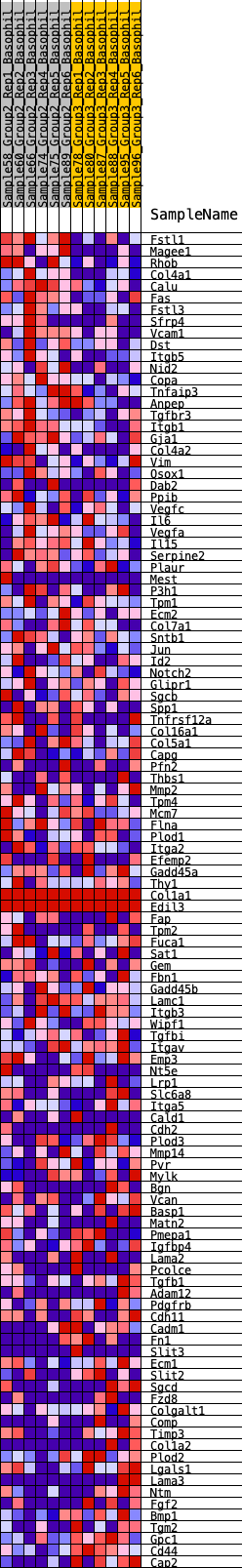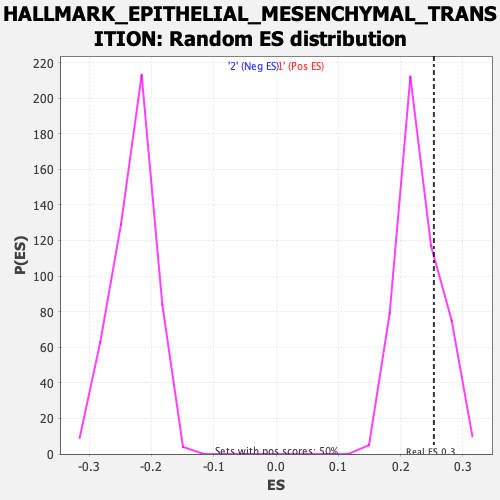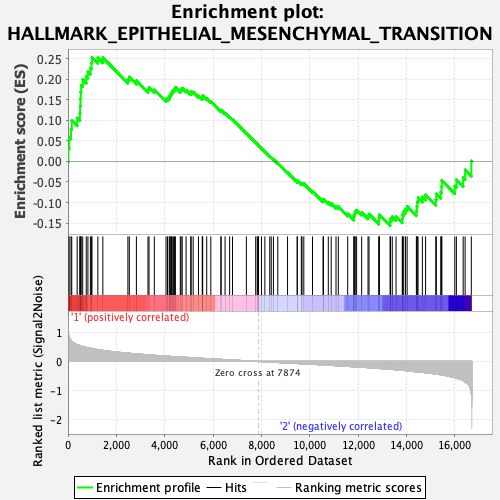
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2534324 |
| Normalized Enrichment Score (NES) | 1.0984116 |
| Nominal p-value | 0.25903615 |
| FDR q-value | 0.6082669 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fstl1 | 17 | 0.989 | 0.0322 | Yes |
| 2 | Magee1 | 48 | 0.847 | 0.0589 | Yes |
| 3 | Rhob | 124 | 0.720 | 0.0786 | Yes |
| 4 | Col4a1 | 155 | 0.688 | 0.1000 | Yes |
| 5 | Calu | 378 | 0.575 | 0.1059 | Yes |
| 6 | Fas | 491 | 0.540 | 0.1173 | Yes |
| 7 | Fstl3 | 501 | 0.538 | 0.1349 | Yes |
| 8 | Sfrp4 | 515 | 0.536 | 0.1521 | Yes |
| 9 | Vcam1 | 527 | 0.532 | 0.1694 | Yes |
| 10 | Dst | 544 | 0.527 | 0.1861 | Yes |
| 11 | Itgb5 | 608 | 0.508 | 0.1994 | Yes |
| 12 | Nid2 | 754 | 0.480 | 0.2068 | Yes |
| 13 | Copa | 823 | 0.469 | 0.2185 | Yes |
| 14 | Tnfaip3 | 926 | 0.452 | 0.2275 | Yes |
| 15 | Anpep | 967 | 0.444 | 0.2400 | Yes |
| 16 | Tgfbr3 | 991 | 0.440 | 0.2534 | Yes |
| 17 | Itgb1 | 1234 | 0.405 | 0.2525 | No |
| 18 | Gja1 | 1441 | 0.379 | 0.2528 | No |
| 19 | Col4a2 | 2477 | 0.285 | 0.2000 | No |
| 20 | Vim | 2535 | 0.279 | 0.2059 | No |
| 21 | Qsox1 | 2828 | 0.258 | 0.1970 | No |
| 22 | Dab2 | 3307 | 0.226 | 0.1758 | No |
| 23 | Ppib | 3350 | 0.224 | 0.1808 | No |
| 24 | Vegfc | 3569 | 0.209 | 0.1747 | No |
| 25 | Il6 | 4054 | 0.182 | 0.1516 | No |
| 26 | Vegfa | 4116 | 0.179 | 0.1539 | No |
| 27 | Il15 | 4185 | 0.175 | 0.1557 | No |
| 28 | Serpine2 | 4210 | 0.173 | 0.1601 | No |
| 29 | Plaur | 4245 | 0.171 | 0.1638 | No |
| 30 | Mest | 4278 | 0.169 | 0.1676 | No |
| 31 | P3h1 | 4311 | 0.169 | 0.1713 | No |
| 32 | Tpm1 | 4367 | 0.167 | 0.1736 | No |
| 33 | Ecm2 | 4402 | 0.165 | 0.1771 | No |
| 34 | Col7a1 | 4441 | 0.163 | 0.1803 | No |
| 35 | Sntb1 | 4644 | 0.152 | 0.1733 | No |
| 36 | Jun | 4674 | 0.151 | 0.1766 | No |
| 37 | Id2 | 4723 | 0.148 | 0.1787 | No |
| 38 | Notch2 | 4883 | 0.139 | 0.1737 | No |
| 39 | Glipr1 | 5077 | 0.130 | 0.1665 | No |
| 40 | Sgcb | 5091 | 0.130 | 0.1701 | No |
| 41 | Spp1 | 5176 | 0.126 | 0.1692 | No |
| 42 | Tnfrsf12a | 5395 | 0.113 | 0.1599 | No |
| 43 | Col16a1 | 5552 | 0.105 | 0.1540 | No |
| 44 | Col5a1 | 5561 | 0.104 | 0.1571 | No |
| 45 | Capg | 5570 | 0.104 | 0.1601 | No |
| 46 | Pfn2 | 5736 | 0.096 | 0.1534 | No |
| 47 | Thbs1 | 5906 | 0.088 | 0.1461 | No |
| 48 | Mmp2 | 6323 | 0.071 | 0.1234 | No |
| 49 | Tpm4 | 6331 | 0.070 | 0.1254 | No |
| 50 | Mcm7 | 6495 | 0.062 | 0.1176 | No |
| 51 | Flna | 6684 | 0.053 | 0.1080 | No |
| 52 | Plod1 | 6802 | 0.047 | 0.1026 | No |
| 53 | Itga2 | 7376 | 0.021 | 0.0687 | No |
| 54 | Efemp2 | 7762 | 0.004 | 0.0457 | No |
| 55 | Gadd45a | 7826 | 0.002 | 0.0419 | No |
| 56 | Thy1 | 7849 | 0.001 | 0.0406 | No |
| 57 | Col1a1 | 7874 | 0.000 | 0.0392 | No |
| 58 | Edil3 | 7880 | 0.000 | 0.0389 | No |
| 59 | Fap | 8001 | -0.005 | 0.0318 | No |
| 60 | Tpm2 | 8141 | -0.011 | 0.0238 | No |
| 61 | Fuca1 | 8343 | -0.021 | 0.0124 | No |
| 62 | Sat1 | 8405 | -0.024 | 0.0095 | No |
| 63 | Gem | 8494 | -0.027 | 0.0051 | No |
| 64 | Fbn1 | 8675 | -0.036 | -0.0046 | No |
| 65 | Gadd45b | 9079 | -0.055 | -0.0270 | No |
| 66 | Lamc1 | 9475 | -0.071 | -0.0484 | No |
| 67 | Itgb3 | 9486 | -0.071 | -0.0466 | No |
| 68 | Wipf1 | 9648 | -0.079 | -0.0537 | No |
| 69 | Tgfbi | 9678 | -0.080 | -0.0527 | No |
| 70 | Itgav | 9752 | -0.083 | -0.0543 | No |
| 71 | Emp3 | 10111 | -0.098 | -0.0726 | No |
| 72 | Nt5e | 10547 | -0.119 | -0.0948 | No |
| 73 | Lrp1 | 10565 | -0.120 | -0.0918 | No |
| 74 | Slc6a8 | 10764 | -0.130 | -0.0994 | No |
| 75 | Itga5 | 10884 | -0.135 | -0.1021 | No |
| 76 | Cald1 | 11083 | -0.144 | -0.1092 | No |
| 77 | Cdh2 | 11178 | -0.149 | -0.1098 | No |
| 78 | Plod3 | 11566 | -0.169 | -0.1275 | No |
| 79 | Mmp14 | 11824 | -0.182 | -0.1369 | No |
| 80 | Pvr | 11825 | -0.182 | -0.1307 | No |
| 81 | Mylk | 11859 | -0.184 | -0.1265 | No |
| 82 | Bgn | 11874 | -0.184 | -0.1212 | No |
| 83 | Vcan | 11935 | -0.188 | -0.1185 | No |
| 84 | Basp1 | 12143 | -0.199 | -0.1243 | No |
| 85 | Matn2 | 12407 | -0.213 | -0.1330 | No |
| 86 | Pmepa1 | 12448 | -0.215 | -0.1282 | No |
| 87 | Igfbp4 | 12856 | -0.239 | -0.1447 | No |
| 88 | Lama2 | 12860 | -0.240 | -0.1368 | No |
| 89 | Pcolce | 12876 | -0.240 | -0.1296 | No |
| 90 | Tgfb1 | 13319 | -0.267 | -0.1473 | No |
| 91 | Adam12 | 13337 | -0.269 | -0.1393 | No |
| 92 | Pdgfrb | 13409 | -0.274 | -0.1343 | No |
| 93 | Cdh11 | 13566 | -0.286 | -0.1341 | No |
| 94 | Cadm1 | 13824 | -0.304 | -0.1394 | No |
| 95 | Fn1 | 13831 | -0.305 | -0.1295 | No |
| 96 | Slit3 | 13881 | -0.309 | -0.1221 | No |
| 97 | Ecm1 | 13954 | -0.313 | -0.1159 | No |
| 98 | Slit2 | 14024 | -0.320 | -0.1093 | No |
| 99 | Sgcd | 14406 | -0.354 | -0.1204 | No |
| 100 | Fzd8 | 14425 | -0.355 | -0.1095 | No |
| 101 | Colgalt1 | 14441 | -0.357 | -0.0984 | No |
| 102 | Comp | 14476 | -0.360 | -0.0884 | No |
| 103 | Timp3 | 14654 | -0.375 | -0.0864 | No |
| 104 | Col1a2 | 14787 | -0.386 | -0.0814 | No |
| 105 | Plod2 | 15210 | -0.430 | -0.0924 | No |
| 106 | Lgals1 | 15231 | -0.433 | -0.0790 | No |
| 107 | Lama3 | 15414 | -0.455 | -0.0747 | No |
| 108 | Ntm | 15451 | -0.460 | -0.0614 | No |
| 109 | Fgf2 | 15457 | -0.461 | -0.0462 | No |
| 110 | Bmp1 | 15995 | -0.556 | -0.0598 | No |
| 111 | Tgm2 | 16061 | -0.576 | -0.0444 | No |
| 112 | Gpc1 | 16341 | -0.655 | -0.0392 | No |
| 113 | Cd44 | 16422 | -0.703 | -0.0204 | No |
| 114 | Cap2 | 16678 | -1.102 | 0.0013 | No |