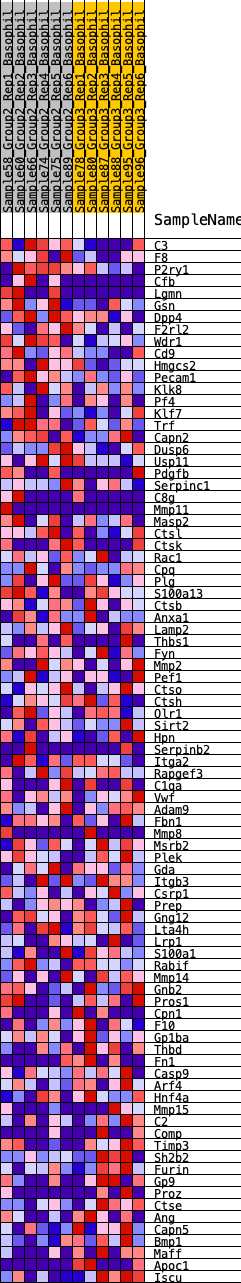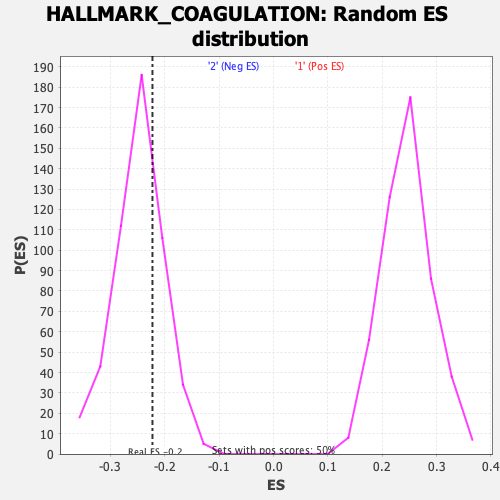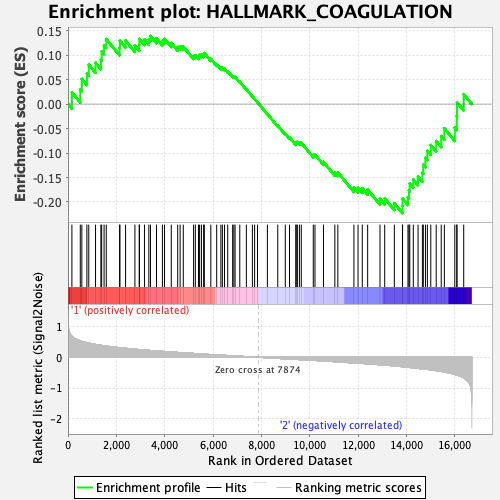
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.22274172 |
| Normalized Enrichment Score (NES) | -0.8990811 |
| Nominal p-value | 0.71626985 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | C3 | 161 | 0.683 | 0.0240 | No |
| 2 | F8 | 508 | 0.537 | 0.0297 | No |
| 3 | P2ry1 | 572 | 0.516 | 0.0514 | No |
| 4 | Cfb | 782 | 0.475 | 0.0622 | No |
| 5 | Lgmn | 859 | 0.463 | 0.0805 | No |
| 6 | Gsn | 1139 | 0.419 | 0.0844 | No |
| 7 | Dpp4 | 1360 | 0.390 | 0.0904 | No |
| 8 | F2rl2 | 1392 | 0.386 | 0.1076 | No |
| 9 | Wdr1 | 1492 | 0.373 | 0.1200 | No |
| 10 | Cd9 | 1582 | 0.364 | 0.1327 | No |
| 11 | Hmgcs2 | 2134 | 0.310 | 0.1148 | No |
| 12 | Pecam1 | 2147 | 0.309 | 0.1293 | No |
| 13 | Klk8 | 2381 | 0.291 | 0.1296 | No |
| 14 | Pf4 | 2767 | 0.262 | 0.1194 | No |
| 15 | Klf7 | 2947 | 0.249 | 0.1209 | No |
| 16 | Trf | 2951 | 0.249 | 0.1330 | No |
| 17 | Capn2 | 3162 | 0.236 | 0.1320 | No |
| 18 | Dusp6 | 3342 | 0.224 | 0.1323 | No |
| 19 | Usp11 | 3410 | 0.221 | 0.1392 | No |
| 20 | Pdgfb | 3660 | 0.204 | 0.1342 | No |
| 21 | Serpinc1 | 3904 | 0.192 | 0.1291 | No |
| 22 | C8g | 3990 | 0.187 | 0.1332 | No |
| 23 | Mmp11 | 4270 | 0.169 | 0.1248 | No |
| 24 | Masp2 | 4536 | 0.158 | 0.1166 | No |
| 25 | Ctsl | 4636 | 0.153 | 0.1182 | No |
| 26 | Ctsk | 4767 | 0.145 | 0.1175 | No |
| 27 | Rac1 | 5194 | 0.125 | 0.0981 | No |
| 28 | Cpq | 5265 | 0.121 | 0.0998 | No |
| 29 | Plg | 5396 | 0.113 | 0.0976 | No |
| 30 | S100a13 | 5442 | 0.111 | 0.1004 | No |
| 31 | Ctsb | 5517 | 0.107 | 0.1012 | No |
| 32 | Anxa1 | 5610 | 0.102 | 0.1007 | No |
| 33 | Lamp2 | 5637 | 0.100 | 0.1041 | No |
| 34 | Thbs1 | 5906 | 0.088 | 0.0923 | No |
| 35 | Fyn | 6149 | 0.077 | 0.0816 | No |
| 36 | Mmp2 | 6323 | 0.071 | 0.0746 | No |
| 37 | Pef1 | 6383 | 0.067 | 0.0744 | No |
| 38 | Ctso | 6471 | 0.063 | 0.0723 | No |
| 39 | Ctsh | 6606 | 0.056 | 0.0670 | No |
| 40 | Olr1 | 6809 | 0.047 | 0.0571 | No |
| 41 | Sirt2 | 6859 | 0.044 | 0.0564 | No |
| 42 | Hpn | 6909 | 0.043 | 0.0555 | No |
| 43 | Serpinb2 | 7104 | 0.034 | 0.0455 | No |
| 44 | Itga2 | 7376 | 0.021 | 0.0302 | No |
| 45 | Rapgef3 | 7625 | 0.011 | 0.0158 | No |
| 46 | C1qa | 7717 | 0.007 | 0.0107 | No |
| 47 | Vwf | 7838 | 0.001 | 0.0035 | No |
| 48 | Adam9 | 8245 | -0.016 | -0.0201 | No |
| 49 | Fbn1 | 8675 | -0.036 | -0.0442 | No |
| 50 | Mmp8 | 8985 | -0.050 | -0.0603 | No |
| 51 | Msrb2 | 9160 | -0.059 | -0.0679 | No |
| 52 | Plek | 9420 | -0.068 | -0.0801 | No |
| 53 | Gda | 9437 | -0.069 | -0.0776 | No |
| 54 | Itgb3 | 9486 | -0.071 | -0.0770 | No |
| 55 | Csrp1 | 9570 | -0.075 | -0.0783 | No |
| 56 | Prep | 9647 | -0.079 | -0.0789 | No |
| 57 | Gng12 | 10143 | -0.099 | -0.1038 | No |
| 58 | Lta4h | 10213 | -0.103 | -0.1029 | No |
| 59 | Lrp1 | 10565 | -0.120 | -0.1181 | No |
| 60 | S100a1 | 11035 | -0.142 | -0.1393 | No |
| 61 | Rabif | 11159 | -0.148 | -0.1394 | No |
| 62 | Mmp14 | 11824 | -0.182 | -0.1704 | No |
| 63 | Gnb2 | 11994 | -0.191 | -0.1712 | No |
| 64 | Pros1 | 12169 | -0.200 | -0.1718 | No |
| 65 | Cpn1 | 12392 | -0.212 | -0.1747 | No |
| 66 | F10 | 12902 | -0.243 | -0.1933 | No |
| 67 | Gp1ba | 13097 | -0.253 | -0.1925 | No |
| 68 | Thbd | 13495 | -0.280 | -0.2026 | No |
| 69 | Fn1 | 13831 | -0.305 | -0.2077 | Yes |
| 70 | Casp9 | 13835 | -0.305 | -0.1928 | Yes |
| 71 | Arf4 | 14059 | -0.323 | -0.1903 | Yes |
| 72 | Hnf4a | 14100 | -0.328 | -0.1765 | Yes |
| 73 | Mmp15 | 14134 | -0.331 | -0.1622 | Yes |
| 74 | C2 | 14279 | -0.343 | -0.1539 | Yes |
| 75 | Comp | 14476 | -0.360 | -0.1480 | Yes |
| 76 | Timp3 | 14654 | -0.375 | -0.1401 | Yes |
| 77 | Sh2b2 | 14693 | -0.380 | -0.1236 | Yes |
| 78 | Furin | 14782 | -0.386 | -0.1099 | Yes |
| 79 | Gp9 | 14861 | -0.392 | -0.0952 | Yes |
| 80 | Proz | 15003 | -0.408 | -0.0836 | Yes |
| 81 | Ctse | 15227 | -0.433 | -0.0756 | Yes |
| 82 | Ang | 15434 | -0.458 | -0.0654 | Yes |
| 83 | Capn5 | 15563 | -0.476 | -0.0496 | Yes |
| 84 | Bmp1 | 15995 | -0.556 | -0.0481 | Yes |
| 85 | Maff | 16078 | -0.579 | -0.0245 | Yes |
| 86 | Apoc1 | 16086 | -0.580 | 0.0037 | Yes |
| 87 | Iscu | 16364 | -0.671 | 0.0202 | Yes |