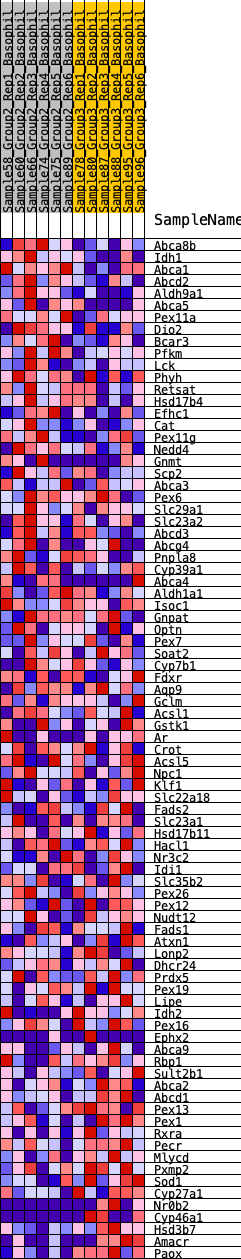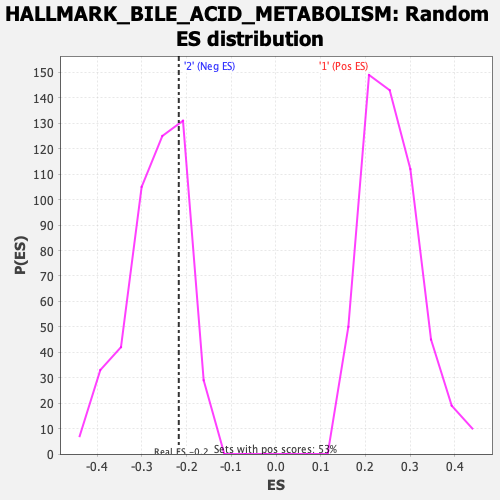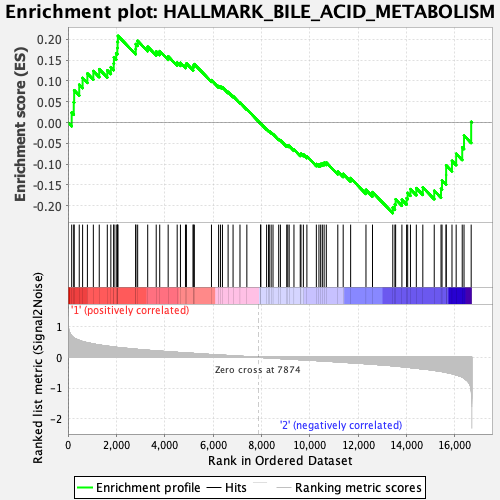
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.21754415 |
| Normalized Enrichment Score (NES) | -0.8161251 |
| Nominal p-value | 0.7584746 |
| FDR q-value | 0.93196744 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca8b | 153 | 0.690 | 0.0237 | Yes |
| 2 | Idh1 | 238 | 0.627 | 0.0486 | Yes |
| 3 | Abca1 | 255 | 0.622 | 0.0774 | Yes |
| 4 | Abcd2 | 465 | 0.547 | 0.0909 | Yes |
| 5 | Aldh9a1 | 604 | 0.510 | 0.1069 | Yes |
| 6 | Abca5 | 802 | 0.473 | 0.1177 | Yes |
| 7 | Pex11a | 1048 | 0.432 | 0.1235 | Yes |
| 8 | Dio2 | 1292 | 0.398 | 0.1279 | Yes |
| 9 | Bcar3 | 1625 | 0.361 | 0.1251 | Yes |
| 10 | Pfkm | 1772 | 0.343 | 0.1327 | Yes |
| 11 | Lck | 1891 | 0.331 | 0.1414 | Yes |
| 12 | Phyh | 1900 | 0.329 | 0.1567 | Yes |
| 13 | Retsat | 1993 | 0.323 | 0.1666 | Yes |
| 14 | Hsd17b4 | 2040 | 0.319 | 0.1790 | Yes |
| 15 | Efhc1 | 2052 | 0.318 | 0.1935 | Yes |
| 16 | Cat | 2065 | 0.317 | 0.2079 | Yes |
| 17 | Pex11g | 2800 | 0.260 | 0.1762 | No |
| 18 | Nedd4 | 2805 | 0.260 | 0.1884 | No |
| 19 | Gnmt | 2881 | 0.254 | 0.1960 | No |
| 20 | Scp2 | 3296 | 0.226 | 0.1819 | No |
| 21 | Abca3 | 3648 | 0.205 | 0.1705 | No |
| 22 | Pex6 | 3793 | 0.198 | 0.1713 | No |
| 23 | Slc29a1 | 4142 | 0.177 | 0.1588 | No |
| 24 | Slc23a2 | 4515 | 0.159 | 0.1440 | No |
| 25 | Abcd3 | 4650 | 0.152 | 0.1432 | No |
| 26 | Abcg4 | 4858 | 0.141 | 0.1374 | No |
| 27 | Pnpla8 | 4897 | 0.139 | 0.1417 | No |
| 28 | Cyp39a1 | 5175 | 0.126 | 0.1311 | No |
| 29 | Abca4 | 5180 | 0.126 | 0.1368 | No |
| 30 | Aldh1a1 | 5225 | 0.123 | 0.1401 | No |
| 31 | Isoc1 | 5933 | 0.087 | 0.1017 | No |
| 32 | Gnpat | 6228 | 0.074 | 0.0875 | No |
| 33 | Optn | 6305 | 0.071 | 0.0864 | No |
| 34 | Pex7 | 6387 | 0.067 | 0.0847 | No |
| 35 | Soat2 | 6621 | 0.056 | 0.0733 | No |
| 36 | Cyp7b1 | 6826 | 0.046 | 0.0632 | No |
| 37 | Fdxr | 7112 | 0.033 | 0.0477 | No |
| 38 | Aqp9 | 7396 | 0.021 | 0.0316 | No |
| 39 | Gclm | 7966 | -0.003 | -0.0025 | No |
| 40 | Acsl1 | 7971 | -0.003 | -0.0026 | No |
| 41 | Gstk1 | 8213 | -0.015 | -0.0164 | No |
| 42 | Ar | 8294 | -0.018 | -0.0203 | No |
| 43 | Crot | 8322 | -0.020 | -0.0210 | No |
| 44 | Acsl5 | 8401 | -0.023 | -0.0246 | No |
| 45 | Npc1 | 8478 | -0.026 | -0.0279 | No |
| 46 | Klf1 | 8713 | -0.037 | -0.0402 | No |
| 47 | Slc22a18 | 8784 | -0.041 | -0.0424 | No |
| 48 | Fads2 | 9045 | -0.053 | -0.0555 | No |
| 49 | Slc23a1 | 9089 | -0.056 | -0.0555 | No |
| 50 | Hsd17b11 | 9146 | -0.058 | -0.0561 | No |
| 51 | Hacl1 | 9344 | -0.065 | -0.0648 | No |
| 52 | Nr3c2 | 9604 | -0.077 | -0.0767 | No |
| 53 | Idi1 | 9634 | -0.078 | -0.0747 | No |
| 54 | Slc35b2 | 9733 | -0.083 | -0.0767 | No |
| 55 | Pex26 | 9879 | -0.089 | -0.0811 | No |
| 56 | Pex12 | 10270 | -0.105 | -0.0996 | No |
| 57 | Nudt12 | 10377 | -0.111 | -0.1007 | No |
| 58 | Fads1 | 10449 | -0.114 | -0.0995 | No |
| 59 | Atxn1 | 10518 | -0.118 | -0.0980 | No |
| 60 | Lonp2 | 10592 | -0.121 | -0.0966 | No |
| 61 | Dhcr24 | 10682 | -0.125 | -0.0960 | No |
| 62 | Prdx5 | 11157 | -0.148 | -0.1175 | No |
| 63 | Pex19 | 11380 | -0.158 | -0.1233 | No |
| 64 | Lipe | 11688 | -0.176 | -0.1333 | No |
| 65 | Idh2 | 12322 | -0.208 | -0.1615 | No |
| 66 | Pex16 | 12592 | -0.222 | -0.1671 | No |
| 67 | Ephx2 | 13431 | -0.276 | -0.2044 | No |
| 68 | Abca9 | 13519 | -0.282 | -0.1961 | No |
| 69 | Rbp1 | 13549 | -0.284 | -0.1843 | No |
| 70 | Sult2b1 | 13808 | -0.303 | -0.1854 | No |
| 71 | Abca2 | 14002 | -0.319 | -0.1818 | No |
| 72 | Abcd1 | 14047 | -0.322 | -0.1690 | No |
| 73 | Pex13 | 14162 | -0.333 | -0.1600 | No |
| 74 | Pex1 | 14404 | -0.354 | -0.1576 | No |
| 75 | Rxra | 14673 | -0.378 | -0.1557 | No |
| 76 | Pecr | 15145 | -0.423 | -0.1638 | No |
| 77 | Mlycd | 15426 | -0.457 | -0.1589 | No |
| 78 | Pxmp2 | 15467 | -0.462 | -0.1392 | No |
| 79 | Sod1 | 15636 | -0.488 | -0.1260 | No |
| 80 | Cyp27a1 | 15638 | -0.488 | -0.1028 | No |
| 81 | Nr0b2 | 15879 | -0.529 | -0.0919 | No |
| 82 | Cyp46a1 | 16052 | -0.572 | -0.0750 | No |
| 83 | Hsd3b7 | 16303 | -0.640 | -0.0595 | No |
| 84 | Amacr | 16377 | -0.679 | -0.0314 | No |
| 85 | Paox | 16673 | -1.062 | 0.0016 | No |