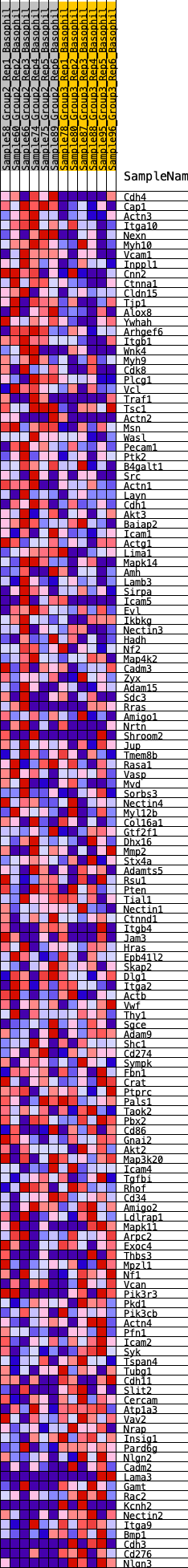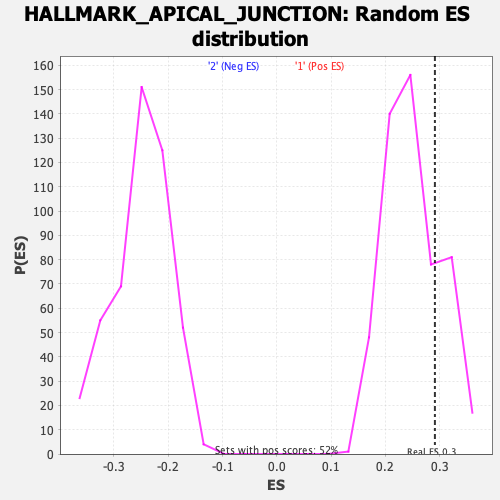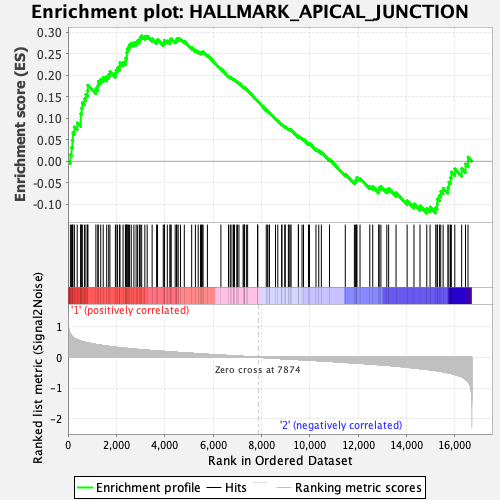
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.29135904 |
| Normalized Enrichment Score (NES) | 1.1698937 |
| Nominal p-value | 0.22264875 |
| FDR q-value | 0.67986655 |
| FWER p-Value | 0.949 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdh4 | 99 | 0.748 | 0.0152 | Yes |
| 2 | Cap1 | 148 | 0.695 | 0.0320 | Yes |
| 3 | Actn3 | 184 | 0.669 | 0.0488 | Yes |
| 4 | Itga10 | 202 | 0.653 | 0.0662 | Yes |
| 5 | Nexn | 264 | 0.618 | 0.0801 | Yes |
| 6 | Myh10 | 381 | 0.575 | 0.0893 | Yes |
| 7 | Vcam1 | 527 | 0.532 | 0.0956 | Yes |
| 8 | Inppl1 | 528 | 0.532 | 0.1107 | Yes |
| 9 | Cnn2 | 565 | 0.519 | 0.1232 | Yes |
| 10 | Ctnna1 | 591 | 0.512 | 0.1362 | Yes |
| 11 | Cldn15 | 680 | 0.494 | 0.1449 | Yes |
| 12 | Tjp1 | 741 | 0.482 | 0.1549 | Yes |
| 13 | Alox8 | 813 | 0.471 | 0.1639 | Yes |
| 14 | Ywhah | 820 | 0.470 | 0.1769 | Yes |
| 15 | Arhgef6 | 1153 | 0.415 | 0.1686 | Yes |
| 16 | Itgb1 | 1234 | 0.405 | 0.1752 | Yes |
| 17 | Wnk4 | 1250 | 0.403 | 0.1857 | Yes |
| 18 | Myh9 | 1353 | 0.392 | 0.1906 | Yes |
| 19 | Cdk8 | 1456 | 0.378 | 0.1952 | Yes |
| 20 | Plcg1 | 1601 | 0.363 | 0.1968 | Yes |
| 21 | Vcl | 1684 | 0.353 | 0.2018 | Yes |
| 22 | Traf1 | 1734 | 0.347 | 0.2087 | Yes |
| 23 | Tsc1 | 1960 | 0.326 | 0.2043 | Yes |
| 24 | Actn2 | 1986 | 0.324 | 0.2120 | Yes |
| 25 | Msn | 2050 | 0.318 | 0.2172 | Yes |
| 26 | Wasl | 2135 | 0.310 | 0.2209 | Yes |
| 27 | Pecam1 | 2147 | 0.309 | 0.2290 | Yes |
| 28 | Ptk2 | 2277 | 0.300 | 0.2296 | Yes |
| 29 | B4galt1 | 2374 | 0.291 | 0.2321 | Yes |
| 30 | Src | 2386 | 0.291 | 0.2397 | Yes |
| 31 | Actn1 | 2432 | 0.287 | 0.2451 | Yes |
| 32 | Layn | 2433 | 0.287 | 0.2532 | Yes |
| 33 | Cdh1 | 2448 | 0.287 | 0.2605 | Yes |
| 34 | Akt3 | 2499 | 0.282 | 0.2654 | Yes |
| 35 | Baiap2 | 2546 | 0.279 | 0.2706 | Yes |
| 36 | Icam1 | 2615 | 0.272 | 0.2742 | Yes |
| 37 | Actg1 | 2725 | 0.265 | 0.2751 | Yes |
| 38 | Lima1 | 2817 | 0.259 | 0.2769 | Yes |
| 39 | Mapk14 | 2879 | 0.254 | 0.2804 | Yes |
| 40 | Amh | 2953 | 0.249 | 0.2831 | Yes |
| 41 | Lamb3 | 2987 | 0.247 | 0.2881 | Yes |
| 42 | Sirpa | 3047 | 0.242 | 0.2914 | Yes |
| 43 | Icam5 | 3178 | 0.234 | 0.2901 | No |
| 44 | Evl | 3274 | 0.229 | 0.2909 | No |
| 45 | Ikbkg | 3479 | 0.216 | 0.2847 | No |
| 46 | Nectin3 | 3664 | 0.203 | 0.2793 | No |
| 47 | Hadh | 3702 | 0.202 | 0.2828 | No |
| 48 | Nf2 | 3939 | 0.190 | 0.2739 | No |
| 49 | Map4k2 | 3981 | 0.187 | 0.2768 | No |
| 50 | Cadm3 | 3993 | 0.186 | 0.2814 | No |
| 51 | Zyx | 4107 | 0.179 | 0.2796 | No |
| 52 | Adam15 | 4218 | 0.173 | 0.2779 | No |
| 53 | Sdc3 | 4223 | 0.172 | 0.2825 | No |
| 54 | Rras | 4268 | 0.169 | 0.2846 | No |
| 55 | Amigo1 | 4432 | 0.164 | 0.2794 | No |
| 56 | Nrtn | 4488 | 0.160 | 0.2807 | No |
| 57 | Shroom2 | 4503 | 0.160 | 0.2843 | No |
| 58 | Jup | 4557 | 0.156 | 0.2855 | No |
| 59 | Tmem8b | 4653 | 0.152 | 0.2841 | No |
| 60 | Rasa1 | 4809 | 0.143 | 0.2788 | No |
| 61 | Vasp | 5114 | 0.129 | 0.2641 | No |
| 62 | Mvd | 5268 | 0.120 | 0.2582 | No |
| 63 | Sorbs3 | 5383 | 0.114 | 0.2546 | No |
| 64 | Nectin4 | 5478 | 0.109 | 0.2520 | No |
| 65 | Myl12b | 5521 | 0.107 | 0.2525 | No |
| 66 | Col16a1 | 5552 | 0.105 | 0.2537 | No |
| 67 | Gtf2f1 | 5586 | 0.103 | 0.2546 | No |
| 68 | Dhx16 | 5765 | 0.095 | 0.2465 | No |
| 69 | Mmp2 | 6323 | 0.071 | 0.2149 | No |
| 70 | Stx4a | 6637 | 0.055 | 0.1975 | No |
| 71 | Adamts5 | 6707 | 0.052 | 0.1948 | No |
| 72 | Rsu1 | 6742 | 0.050 | 0.1942 | No |
| 73 | Pten | 6829 | 0.046 | 0.1903 | No |
| 74 | Tial1 | 6842 | 0.045 | 0.1908 | No |
| 75 | Nectin1 | 6886 | 0.043 | 0.1895 | No |
| 76 | Ctnnd1 | 6981 | 0.039 | 0.1849 | No |
| 77 | Itgb4 | 7004 | 0.038 | 0.1846 | No |
| 78 | Jam3 | 7069 | 0.035 | 0.1818 | No |
| 79 | Hras | 7241 | 0.027 | 0.1722 | No |
| 80 | Epb41l2 | 7280 | 0.026 | 0.1707 | No |
| 81 | Skap2 | 7282 | 0.026 | 0.1713 | No |
| 82 | Dlg1 | 7297 | 0.025 | 0.1712 | No |
| 83 | Itga2 | 7376 | 0.021 | 0.1671 | No |
| 84 | Actb | 7428 | 0.019 | 0.1646 | No |
| 85 | Vwf | 7838 | 0.001 | 0.1399 | No |
| 86 | Thy1 | 7849 | 0.001 | 0.1393 | No |
| 87 | Sgce | 8198 | -0.014 | 0.1187 | No |
| 88 | Adam9 | 8245 | -0.016 | 0.1164 | No |
| 89 | Shc1 | 8316 | -0.019 | 0.1127 | No |
| 90 | Cd274 | 8330 | -0.020 | 0.1125 | No |
| 91 | Sympk | 8581 | -0.031 | 0.0983 | No |
| 92 | Fbn1 | 8675 | -0.036 | 0.0937 | No |
| 93 | Crat | 8835 | -0.044 | 0.0853 | No |
| 94 | Ptprc | 8848 | -0.045 | 0.0858 | No |
| 95 | Pals1 | 8963 | -0.049 | 0.0803 | No |
| 96 | Taok2 | 8984 | -0.050 | 0.0806 | No |
| 97 | Pbx2 | 9113 | -0.057 | 0.0744 | No |
| 98 | Cd86 | 9140 | -0.058 | 0.0745 | No |
| 99 | Gnai2 | 9185 | -0.060 | 0.0735 | No |
| 100 | Akt2 | 9225 | -0.062 | 0.0729 | No |
| 101 | Map3k20 | 9524 | -0.073 | 0.0570 | No |
| 102 | Icam4 | 9533 | -0.073 | 0.0586 | No |
| 103 | Tgfbi | 9678 | -0.080 | 0.0522 | No |
| 104 | Rhof | 9739 | -0.083 | 0.0509 | No |
| 105 | Cd34 | 9946 | -0.092 | 0.0410 | No |
| 106 | Amigo2 | 9986 | -0.094 | 0.0413 | No |
| 107 | Ldlrap1 | 10254 | -0.104 | 0.0282 | No |
| 108 | Mapk11 | 10370 | -0.111 | 0.0243 | No |
| 109 | Arpc2 | 10477 | -0.116 | 0.0212 | No |
| 110 | Exoc4 | 10814 | -0.131 | 0.0046 | No |
| 111 | Thbs3 | 11466 | -0.163 | -0.0301 | No |
| 112 | Mpzl1 | 11843 | -0.183 | -0.0476 | No |
| 113 | Nf1 | 11886 | -0.185 | -0.0449 | No |
| 114 | Vcan | 11935 | -0.188 | -0.0425 | No |
| 115 | Pik3r3 | 11948 | -0.188 | -0.0379 | No |
| 116 | Pkd1 | 12076 | -0.195 | -0.0400 | No |
| 117 | Pik3cb | 12480 | -0.217 | -0.0582 | No |
| 118 | Actn4 | 12602 | -0.223 | -0.0592 | No |
| 119 | Pfn1 | 12849 | -0.239 | -0.0673 | No |
| 120 | Icam2 | 12867 | -0.240 | -0.0616 | No |
| 121 | Syk | 12940 | -0.245 | -0.0590 | No |
| 122 | Tspan4 | 13184 | -0.260 | -0.0663 | No |
| 123 | Tubg1 | 13260 | -0.264 | -0.0633 | No |
| 124 | Cdh11 | 13566 | -0.286 | -0.0737 | No |
| 125 | Slit2 | 14024 | -0.320 | -0.0922 | No |
| 126 | Cercam | 14307 | -0.346 | -0.0994 | No |
| 127 | Atp1a3 | 14557 | -0.368 | -0.1041 | No |
| 128 | Vav2 | 14837 | -0.389 | -0.1099 | No |
| 129 | Nrap | 14972 | -0.403 | -0.1066 | No |
| 130 | Insig1 | 15207 | -0.430 | -0.1085 | No |
| 131 | Pard6g | 15263 | -0.437 | -0.0995 | No |
| 132 | Nlgn2 | 15274 | -0.438 | -0.0877 | No |
| 133 | Cadm2 | 15360 | -0.450 | -0.0801 | No |
| 134 | Lama3 | 15414 | -0.455 | -0.0704 | No |
| 135 | Gamt | 15513 | -0.467 | -0.0631 | No |
| 136 | Rac2 | 15709 | -0.502 | -0.0606 | No |
| 137 | Kcnh2 | 15751 | -0.508 | -0.0487 | No |
| 138 | Nectin2 | 15820 | -0.521 | -0.0381 | No |
| 139 | Itga9 | 15852 | -0.526 | -0.0251 | No |
| 140 | Bmp1 | 15995 | -0.556 | -0.0179 | No |
| 141 | Cdh3 | 16279 | -0.635 | -0.0170 | No |
| 142 | Cd276 | 16438 | -0.710 | -0.0065 | No |
| 143 | Nlgn3 | 16541 | -0.783 | 0.0095 | No |