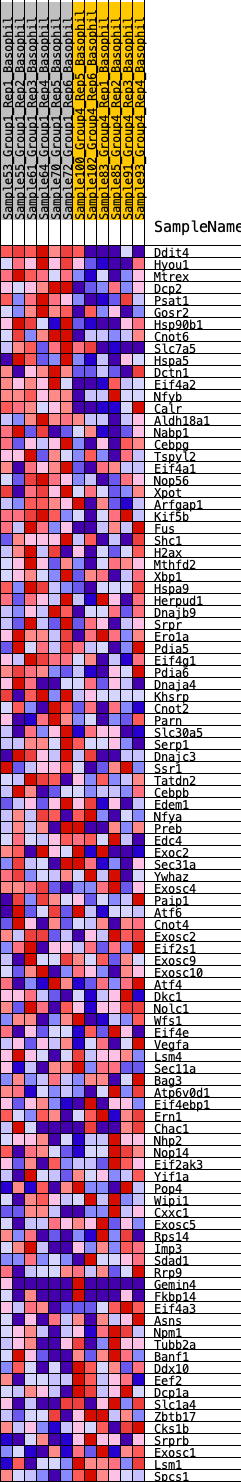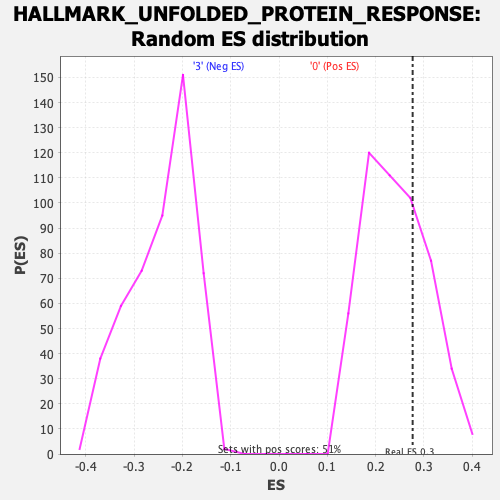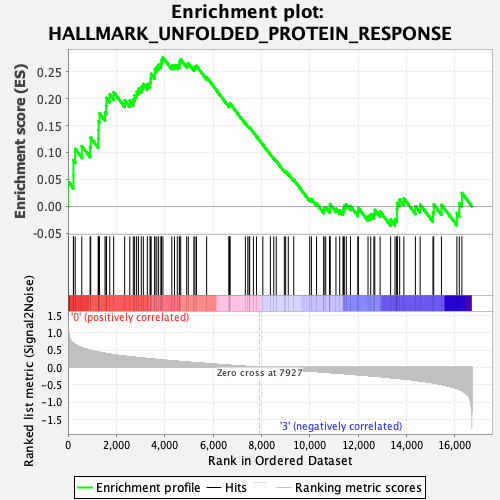
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.27683896 |
| Normalized Enrichment Score (NES) | 1.1401786 |
| Nominal p-value | 0.28543308 |
| FDR q-value | 0.7555049 |
| FWER p-Value | 0.988 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ddit4 | 6 | 1.210 | 0.0460 | Yes |
| 2 | Hyou1 | 225 | 0.692 | 0.0594 | Yes |
| 3 | Mtrex | 226 | 0.690 | 0.0858 | Yes |
| 4 | Dcp2 | 297 | 0.657 | 0.1067 | Yes |
| 5 | Psat1 | 570 | 0.567 | 0.1121 | Yes |
| 6 | Gosr2 | 914 | 0.495 | 0.1103 | Yes |
| 7 | Hsp90b1 | 940 | 0.490 | 0.1276 | Yes |
| 8 | Cnot6 | 1250 | 0.439 | 0.1258 | Yes |
| 9 | Slc7a5 | 1257 | 0.438 | 0.1423 | Yes |
| 10 | Hspa5 | 1270 | 0.437 | 0.1583 | Yes |
| 11 | Dctn1 | 1304 | 0.433 | 0.1729 | Yes |
| 12 | Eif4a2 | 1531 | 0.403 | 0.1747 | Yes |
| 13 | Nfyb | 1577 | 0.398 | 0.1872 | Yes |
| 14 | Calr | 1594 | 0.396 | 0.2014 | Yes |
| 15 | Aldh18a1 | 1726 | 0.380 | 0.2081 | Yes |
| 16 | Nabp1 | 1889 | 0.362 | 0.2122 | Yes |
| 17 | Cebpg | 2343 | 0.323 | 0.1973 | Yes |
| 18 | Tspyl2 | 2554 | 0.305 | 0.1963 | Yes |
| 19 | Eif4a1 | 2700 | 0.294 | 0.1988 | Yes |
| 20 | Nop56 | 2756 | 0.289 | 0.2066 | Yes |
| 21 | Xpot | 2837 | 0.285 | 0.2127 | Yes |
| 22 | Arfgap1 | 2912 | 0.279 | 0.2189 | Yes |
| 23 | Kif5b | 3030 | 0.271 | 0.2223 | Yes |
| 24 | Fus | 3114 | 0.265 | 0.2274 | Yes |
| 25 | Shc1 | 3281 | 0.254 | 0.2272 | Yes |
| 26 | H2ax | 3392 | 0.248 | 0.2300 | Yes |
| 27 | Mthfd2 | 3418 | 0.245 | 0.2379 | Yes |
| 28 | Xbp1 | 3435 | 0.244 | 0.2463 | Yes |
| 29 | Hspa9 | 3589 | 0.234 | 0.2461 | Yes |
| 30 | Herpud1 | 3591 | 0.234 | 0.2549 | Yes |
| 31 | Dnajb9 | 3671 | 0.227 | 0.2589 | Yes |
| 32 | Srpr | 3746 | 0.221 | 0.2629 | Yes |
| 33 | Ero1a | 3842 | 0.216 | 0.2654 | Yes |
| 34 | Pdia5 | 3864 | 0.214 | 0.2724 | Yes |
| 35 | Eif4g1 | 3925 | 0.211 | 0.2768 | Yes |
| 36 | Pdia6 | 4294 | 0.189 | 0.2619 | No |
| 37 | Dnaja4 | 4403 | 0.182 | 0.2624 | No |
| 38 | Khsrp | 4520 | 0.175 | 0.2621 | No |
| 39 | Cnot2 | 4604 | 0.170 | 0.2636 | No |
| 40 | Parn | 4615 | 0.169 | 0.2695 | No |
| 41 | Slc30a5 | 4662 | 0.167 | 0.2731 | No |
| 42 | Serp1 | 4908 | 0.153 | 0.2642 | No |
| 43 | Dnajc3 | 4975 | 0.150 | 0.2659 | No |
| 44 | Ssr1 | 5206 | 0.136 | 0.2573 | No |
| 45 | Tatdn2 | 5250 | 0.134 | 0.2598 | No |
| 46 | Cebpb | 5314 | 0.130 | 0.2610 | No |
| 47 | Edem1 | 5734 | 0.111 | 0.2401 | No |
| 48 | Nfya | 6656 | 0.061 | 0.1869 | No |
| 49 | Preb | 6664 | 0.061 | 0.1888 | No |
| 50 | Edc4 | 6682 | 0.060 | 0.1901 | No |
| 51 | Exoc2 | 6704 | 0.059 | 0.1911 | No |
| 52 | Sec31a | 7334 | 0.029 | 0.1543 | No |
| 53 | Ywhaz | 7436 | 0.024 | 0.1492 | No |
| 54 | Exosc4 | 7494 | 0.021 | 0.1466 | No |
| 55 | Paip1 | 7512 | 0.020 | 0.1463 | No |
| 56 | Atf6 | 7667 | 0.012 | 0.1375 | No |
| 57 | Cnot4 | 7793 | 0.006 | 0.1302 | No |
| 58 | Exosc2 | 8057 | -0.004 | 0.1145 | No |
| 59 | Eif2s1 | 8369 | -0.019 | 0.0965 | No |
| 60 | Exosc9 | 8509 | -0.025 | 0.0891 | No |
| 61 | Exosc10 | 8611 | -0.030 | 0.0842 | No |
| 62 | Atf4 | 8946 | -0.048 | 0.0659 | No |
| 63 | Dkc1 | 9001 | -0.051 | 0.0646 | No |
| 64 | Nolc1 | 9110 | -0.057 | 0.0603 | No |
| 65 | Wfs1 | 9335 | -0.069 | 0.0494 | No |
| 66 | Eif4e | 9995 | -0.102 | 0.0136 | No |
| 67 | Vegfa | 10066 | -0.105 | 0.0134 | No |
| 68 | Lsm4 | 10279 | -0.116 | 0.0051 | No |
| 69 | Sec11a | 10579 | -0.133 | -0.0078 | No |
| 70 | Bag3 | 10584 | -0.133 | -0.0030 | No |
| 71 | Atp6v0d1 | 10658 | -0.137 | -0.0021 | No |
| 72 | Eif4ebp1 | 10817 | -0.145 | -0.0061 | No |
| 73 | Ern1 | 10830 | -0.146 | -0.0012 | No |
| 74 | Chac1 | 10835 | -0.146 | 0.0041 | No |
| 75 | Nhp2 | 11081 | -0.157 | -0.0046 | No |
| 76 | Nop14 | 11234 | -0.166 | -0.0074 | No |
| 77 | Eif2ak3 | 11371 | -0.174 | -0.0089 | No |
| 78 | Yif1a | 11396 | -0.176 | -0.0037 | No |
| 79 | Pop4 | 11425 | -0.177 | 0.0014 | No |
| 80 | Wipi1 | 11508 | -0.182 | 0.0035 | No |
| 81 | Cxxc1 | 11681 | -0.190 | 0.0004 | No |
| 82 | Exosc5 | 11985 | -0.209 | -0.0099 | No |
| 83 | Rps14 | 12008 | -0.210 | -0.0031 | No |
| 84 | Imp3 | 12407 | -0.233 | -0.0182 | No |
| 85 | Sdad1 | 12518 | -0.241 | -0.0156 | No |
| 86 | Rrp9 | 12648 | -0.246 | -0.0139 | No |
| 87 | Gemin4 | 12688 | -0.248 | -0.0068 | No |
| 88 | Fkbp14 | 12907 | -0.261 | -0.0099 | No |
| 89 | Eif4a3 | 13341 | -0.291 | -0.0248 | No |
| 90 | Asns | 13524 | -0.305 | -0.0241 | No |
| 91 | Npm1 | 13601 | -0.310 | -0.0168 | No |
| 92 | Tubb2a | 13602 | -0.310 | -0.0049 | No |
| 93 | Banf1 | 13615 | -0.311 | 0.0063 | No |
| 94 | Ddx10 | 13716 | -0.317 | 0.0124 | No |
| 95 | Eef2 | 13889 | -0.328 | 0.0146 | No |
| 96 | Dcp1a | 14366 | -0.368 | -0.0000 | No |
| 97 | Slc1a4 | 14562 | -0.388 | 0.0031 | No |
| 98 | Zbtb17 | 15093 | -0.442 | -0.0119 | No |
| 99 | Cks1b | 15123 | -0.446 | 0.0034 | No |
| 100 | Srprb | 15444 | -0.487 | 0.0028 | No |
| 101 | Exosc1 | 16082 | -0.602 | -0.0125 | No |
| 102 | Lsm1 | 16182 | -0.631 | 0.0057 | No |
| 103 | Spcs1 | 16289 | -0.663 | 0.0247 | No |