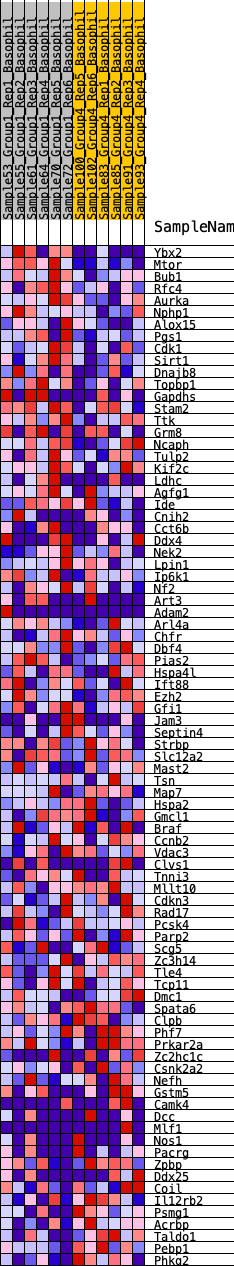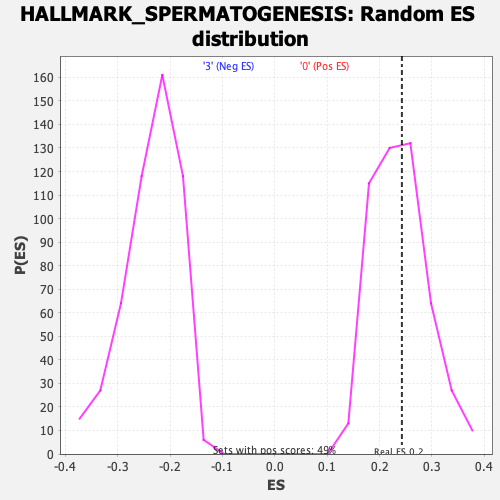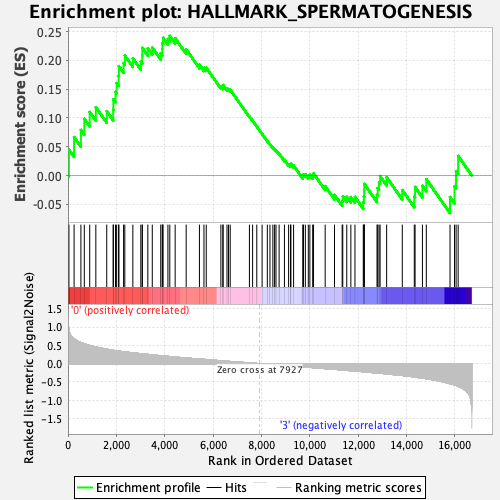
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.242697 |
| Normalized Enrichment Score (NES) | 1.018359 |
| Nominal p-value | 0.43584523 |
| FDR q-value | 0.7908866 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ybx2 | 38 | 0.925 | 0.0450 | Yes |
| 2 | Mtor | 252 | 0.677 | 0.0668 | Yes |
| 3 | Bub1 | 534 | 0.574 | 0.0792 | Yes |
| 4 | Rfc4 | 674 | 0.541 | 0.0985 | Yes |
| 5 | Aurka | 898 | 0.497 | 0.1105 | Yes |
| 6 | Nphp1 | 1151 | 0.456 | 0.1187 | Yes |
| 7 | Alox15 | 1602 | 0.395 | 0.1118 | Yes |
| 8 | Pgs1 | 1868 | 0.364 | 0.1145 | Yes |
| 9 | Cdk1 | 1876 | 0.364 | 0.1326 | Yes |
| 10 | Sirt1 | 1965 | 0.354 | 0.1454 | Yes |
| 11 | Dnajb8 | 2011 | 0.349 | 0.1606 | Yes |
| 12 | Topbp1 | 2094 | 0.343 | 0.1732 | Yes |
| 13 | Gapdhs | 2104 | 0.342 | 0.1901 | Yes |
| 14 | Stam2 | 2299 | 0.327 | 0.1951 | Yes |
| 15 | Ttk | 2349 | 0.323 | 0.2087 | Yes |
| 16 | Grm8 | 2682 | 0.296 | 0.2038 | Yes |
| 17 | Ncaph | 3011 | 0.272 | 0.1980 | Yes |
| 18 | Tulp2 | 3071 | 0.269 | 0.2082 | Yes |
| 19 | Kif2c | 3073 | 0.269 | 0.2219 | Yes |
| 20 | Ldhc | 3301 | 0.252 | 0.2212 | Yes |
| 21 | Agfg1 | 3485 | 0.241 | 0.2225 | Yes |
| 22 | Ide | 3835 | 0.216 | 0.2125 | Yes |
| 23 | Cnih2 | 3900 | 0.212 | 0.2195 | Yes |
| 24 | Cct6b | 3903 | 0.212 | 0.2302 | Yes |
| 25 | Ddx4 | 3932 | 0.210 | 0.2393 | Yes |
| 26 | Nek2 | 4132 | 0.199 | 0.2374 | Yes |
| 27 | Lpin1 | 4210 | 0.194 | 0.2427 | Yes |
| 28 | Ip6k1 | 4431 | 0.180 | 0.2387 | No |
| 29 | Nf2 | 4887 | 0.155 | 0.2192 | No |
| 30 | Art3 | 5435 | 0.124 | 0.1926 | No |
| 31 | Adam2 | 5622 | 0.115 | 0.1873 | No |
| 32 | Arl4a | 5717 | 0.113 | 0.1873 | No |
| 33 | Chfr | 6322 | 0.078 | 0.1550 | No |
| 34 | Dbf4 | 6406 | 0.074 | 0.1538 | No |
| 35 | Pias2 | 6412 | 0.074 | 0.1573 | No |
| 36 | Hspa4l | 6575 | 0.066 | 0.1509 | No |
| 37 | Ift88 | 6639 | 0.062 | 0.1503 | No |
| 38 | Ezh2 | 6708 | 0.059 | 0.1492 | No |
| 39 | Gfi1 | 7502 | 0.021 | 0.1025 | No |
| 40 | Jam3 | 7634 | 0.014 | 0.0954 | No |
| 41 | Septin4 | 7807 | 0.006 | 0.0853 | No |
| 42 | Strbp | 8027 | -0.003 | 0.0723 | No |
| 43 | Slc12a2 | 8243 | -0.013 | 0.0600 | No |
| 44 | Mast2 | 8351 | -0.018 | 0.0545 | No |
| 45 | Tsn | 8472 | -0.023 | 0.0485 | No |
| 46 | Map7 | 8543 | -0.026 | 0.0456 | No |
| 47 | Hspa2 | 8601 | -0.030 | 0.0437 | No |
| 48 | Gmcl1 | 8730 | -0.037 | 0.0379 | No |
| 49 | Braf | 8950 | -0.048 | 0.0272 | No |
| 50 | Ccnb2 | 9123 | -0.057 | 0.0197 | No |
| 51 | Vdac3 | 9201 | -0.062 | 0.0182 | No |
| 52 | Clvs1 | 9218 | -0.062 | 0.0205 | No |
| 53 | Tnni3 | 9329 | -0.068 | 0.0173 | No |
| 54 | Mllt10 | 9708 | -0.089 | -0.0009 | No |
| 55 | Cdkn3 | 9738 | -0.090 | 0.0020 | No |
| 56 | Rad17 | 9815 | -0.094 | 0.0022 | No |
| 57 | Pcsk4 | 9938 | -0.099 | -0.0000 | No |
| 58 | Parp2 | 10002 | -0.102 | 0.0014 | No |
| 59 | Scg5 | 10119 | -0.108 | -0.0001 | No |
| 60 | Zc3h14 | 10153 | -0.110 | 0.0036 | No |
| 61 | Tle4 | 10635 | -0.136 | -0.0185 | No |
| 62 | Tcp11 | 11021 | -0.154 | -0.0338 | No |
| 63 | Dmc1 | 11337 | -0.172 | -0.0439 | No |
| 64 | Spata6 | 11363 | -0.174 | -0.0365 | No |
| 65 | Clpb | 11528 | -0.183 | -0.0370 | No |
| 66 | Phf7 | 11697 | -0.191 | -0.0374 | No |
| 67 | Prkar2a | 11867 | -0.202 | -0.0372 | No |
| 68 | Zc2hc1c | 12211 | -0.222 | -0.0465 | No |
| 69 | Csnk2a2 | 12235 | -0.223 | -0.0365 | No |
| 70 | Nefh | 12250 | -0.223 | -0.0260 | No |
| 71 | Gstm5 | 12252 | -0.224 | -0.0146 | No |
| 72 | Camk4 | 12779 | -0.255 | -0.0332 | No |
| 73 | Dcc | 12808 | -0.256 | -0.0218 | No |
| 74 | Mlf1 | 12863 | -0.258 | -0.0118 | No |
| 75 | Nos1 | 12906 | -0.261 | -0.0010 | No |
| 76 | Pacrg | 13177 | -0.279 | -0.0030 | No |
| 77 | Zpbp | 13824 | -0.323 | -0.0254 | No |
| 78 | Ddx25 | 14320 | -0.362 | -0.0367 | No |
| 79 | Coil | 14353 | -0.366 | -0.0199 | No |
| 80 | Il12rb2 | 14657 | -0.397 | -0.0178 | No |
| 81 | Psmg1 | 14817 | -0.410 | -0.0064 | No |
| 82 | Acrbp | 15798 | -0.549 | -0.0373 | No |
| 83 | Taldo1 | 15984 | -0.579 | -0.0189 | No |
| 84 | Pebp1 | 16049 | -0.595 | 0.0077 | No |
| 85 | Phkg2 | 16134 | -0.613 | 0.0340 | No |