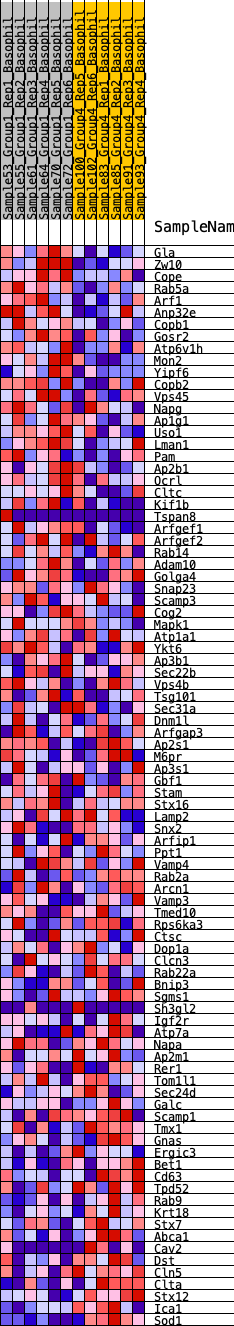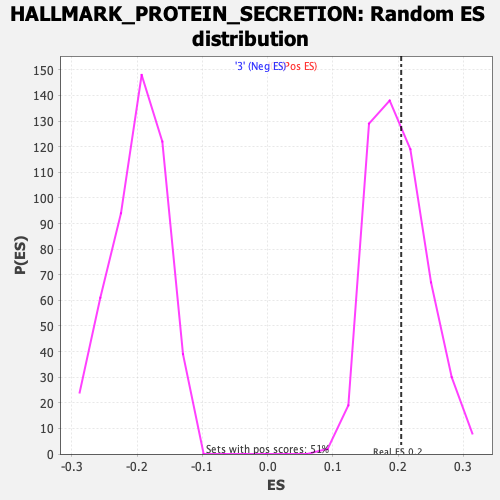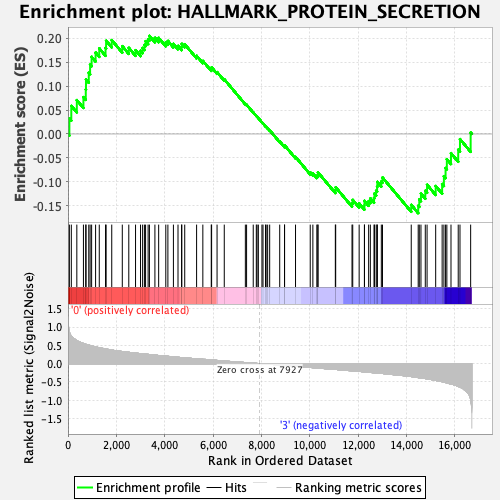
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.2048132 |
| Normalized Enrichment Score (NES) | 1.0281489 |
| Nominal p-value | 0.42773438 |
| FDR q-value | 0.84160155 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gla | 57 | 0.877 | 0.0326 | Yes |
| 2 | Zw10 | 137 | 0.748 | 0.0586 | Yes |
| 3 | Cope | 365 | 0.628 | 0.0707 | Yes |
| 4 | Rab5a | 638 | 0.550 | 0.0769 | Yes |
| 5 | Arf1 | 735 | 0.528 | 0.0928 | Yes |
| 6 | Anp32e | 744 | 0.527 | 0.1140 | Yes |
| 7 | Copb1 | 859 | 0.504 | 0.1278 | Yes |
| 8 | Gosr2 | 914 | 0.495 | 0.1449 | Yes |
| 9 | Atp6v1h | 977 | 0.483 | 0.1610 | Yes |
| 10 | Mon2 | 1141 | 0.457 | 0.1700 | Yes |
| 11 | Yipf6 | 1293 | 0.434 | 0.1787 | Yes |
| 12 | Copb2 | 1552 | 0.402 | 0.1797 | Yes |
| 13 | Vps45 | 1576 | 0.398 | 0.1947 | Yes |
| 14 | Napg | 1809 | 0.372 | 0.1960 | Yes |
| 15 | Ap1g1 | 2245 | 0.329 | 0.1833 | Yes |
| 16 | Uso1 | 2514 | 0.309 | 0.1798 | Yes |
| 17 | Lman1 | 2793 | 0.286 | 0.1748 | Yes |
| 18 | Pam | 2999 | 0.273 | 0.1737 | Yes |
| 19 | Ap2b1 | 3086 | 0.268 | 0.1796 | Yes |
| 20 | Ocrl | 3160 | 0.262 | 0.1859 | Yes |
| 21 | Cltc | 3208 | 0.258 | 0.1937 | Yes |
| 22 | Kif1b | 3313 | 0.252 | 0.1978 | Yes |
| 23 | Tspan8 | 3367 | 0.249 | 0.2048 | Yes |
| 24 | Arfgef1 | 3594 | 0.233 | 0.2008 | No |
| 25 | Arfgef2 | 3749 | 0.221 | 0.2006 | No |
| 26 | Rab14 | 4044 | 0.204 | 0.1913 | No |
| 27 | Adam10 | 4131 | 0.199 | 0.1943 | No |
| 28 | Golga4 | 4359 | 0.184 | 0.1882 | No |
| 29 | Snap23 | 4549 | 0.174 | 0.1839 | No |
| 30 | Scamp3 | 4698 | 0.165 | 0.1818 | No |
| 31 | Cog2 | 4705 | 0.164 | 0.1882 | No |
| 32 | Mapk1 | 4827 | 0.157 | 0.1873 | No |
| 33 | Atp1a1 | 5318 | 0.130 | 0.1632 | No |
| 34 | Ykt6 | 5576 | 0.117 | 0.1525 | No |
| 35 | Ap3b1 | 5924 | 0.101 | 0.1358 | No |
| 36 | Sec22b | 5939 | 0.100 | 0.1390 | No |
| 37 | Vps4b | 6164 | 0.087 | 0.1291 | No |
| 38 | Tsg101 | 6462 | 0.071 | 0.1142 | No |
| 39 | Sec31a | 7334 | 0.029 | 0.0630 | No |
| 40 | Dnm1l | 7385 | 0.027 | 0.0610 | No |
| 41 | Arfgap3 | 7660 | 0.013 | 0.0451 | No |
| 42 | Ap2s1 | 7787 | 0.007 | 0.0378 | No |
| 43 | M6pr | 7818 | 0.005 | 0.0362 | No |
| 44 | Ap3s1 | 7872 | 0.003 | 0.0331 | No |
| 45 | Gbf1 | 8020 | -0.003 | 0.0244 | No |
| 46 | Stam | 8064 | -0.004 | 0.0219 | No |
| 47 | Stx16 | 8168 | -0.009 | 0.0161 | No |
| 48 | Lamp2 | 8193 | -0.011 | 0.0151 | No |
| 49 | Snx2 | 8256 | -0.014 | 0.0120 | No |
| 50 | Arfip1 | 8340 | -0.018 | 0.0077 | No |
| 51 | Ppt1 | 8755 | -0.038 | -0.0157 | No |
| 52 | Vamp4 | 8954 | -0.048 | -0.0256 | No |
| 53 | Rab2a | 8956 | -0.049 | -0.0237 | No |
| 54 | Arcn1 | 9409 | -0.074 | -0.0478 | No |
| 55 | Vamp3 | 10017 | -0.103 | -0.0802 | No |
| 56 | Tmed10 | 10126 | -0.108 | -0.0822 | No |
| 57 | Rps6ka3 | 10290 | -0.117 | -0.0872 | No |
| 58 | Ctsc | 10334 | -0.119 | -0.0849 | No |
| 59 | Dop1a | 10336 | -0.120 | -0.0801 | No |
| 60 | Clcn3 | 11055 | -0.156 | -0.1169 | No |
| 61 | Rab22a | 11070 | -0.157 | -0.1113 | No |
| 62 | Bnip3 | 11745 | -0.194 | -0.1439 | No |
| 63 | Sgms1 | 11775 | -0.196 | -0.1376 | No |
| 64 | Sh3gl2 | 12040 | -0.211 | -0.1448 | No |
| 65 | Igf2r | 12258 | -0.224 | -0.1487 | No |
| 66 | Atp7a | 12260 | -0.224 | -0.1395 | No |
| 67 | Napa | 12433 | -0.235 | -0.1402 | No |
| 68 | Ap2m1 | 12503 | -0.240 | -0.1345 | No |
| 69 | Rer1 | 12656 | -0.246 | -0.1335 | No |
| 70 | Tom1l1 | 12672 | -0.248 | -0.1243 | No |
| 71 | Sec24d | 12744 | -0.252 | -0.1182 | No |
| 72 | Galc | 12783 | -0.255 | -0.1100 | No |
| 73 | Scamp1 | 12792 | -0.256 | -0.1000 | No |
| 74 | Tmx1 | 12957 | -0.264 | -0.0990 | No |
| 75 | Gnas | 13005 | -0.267 | -0.0909 | No |
| 76 | Ergic3 | 14194 | -0.351 | -0.1480 | No |
| 77 | Bet1 | 14482 | -0.380 | -0.1496 | No |
| 78 | Cd63 | 14527 | -0.385 | -0.1365 | No |
| 79 | Tpd52 | 14596 | -0.391 | -0.1245 | No |
| 80 | Rab9 | 14775 | -0.407 | -0.1185 | No |
| 81 | Krt18 | 14849 | -0.412 | -0.1060 | No |
| 82 | Stx7 | 15205 | -0.457 | -0.1086 | No |
| 83 | Abca1 | 15472 | -0.492 | -0.1044 | No |
| 84 | Cav2 | 15545 | -0.503 | -0.0881 | No |
| 85 | Dst | 15614 | -0.515 | -0.0710 | No |
| 86 | Cln5 | 15667 | -0.525 | -0.0526 | No |
| 87 | Clta | 15838 | -0.554 | -0.0401 | No |
| 88 | Stx12 | 16137 | -0.615 | -0.0327 | No |
| 89 | Ica1 | 16210 | -0.640 | -0.0108 | No |
| 90 | Sod1 | 16651 | -0.977 | 0.0029 | No |