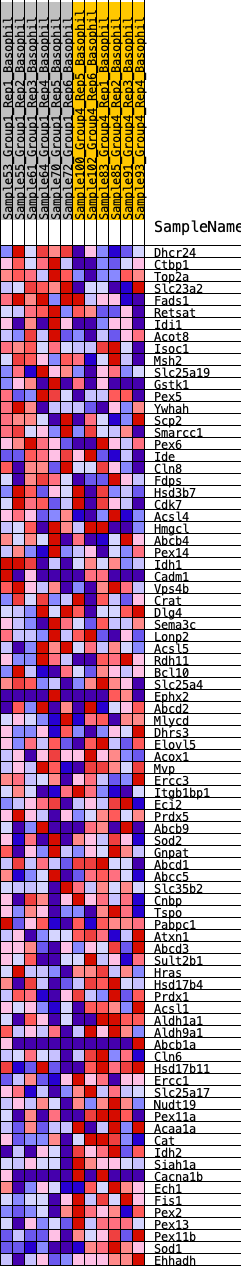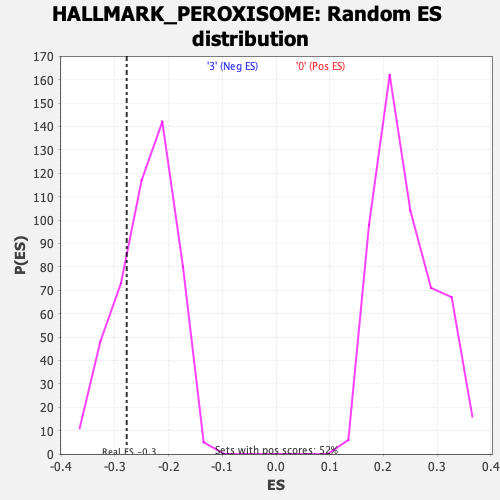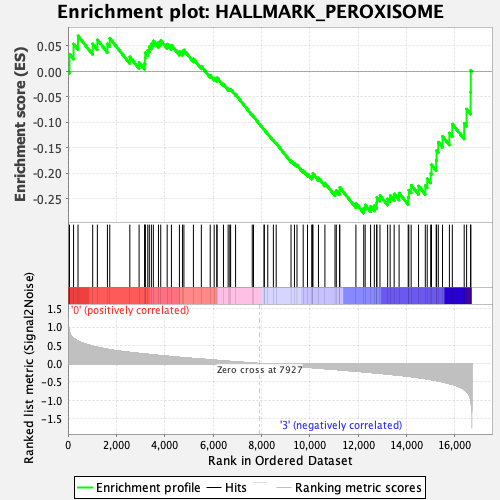
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.27786148 |
| Normalized Enrichment Score (NES) | -1.1516258 |
| Nominal p-value | 0.24789916 |
| FDR q-value | 0.49445686 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 61 | 0.862 | 0.0341 | No |
| 2 | Ctbp1 | 230 | 0.688 | 0.0541 | No |
| 3 | Top2a | 414 | 0.610 | 0.0699 | No |
| 4 | Slc23a2 | 1021 | 0.475 | 0.0542 | No |
| 5 | Fads1 | 1214 | 0.446 | 0.0622 | No |
| 6 | Retsat | 1630 | 0.392 | 0.0544 | No |
| 7 | Idi1 | 1727 | 0.380 | 0.0653 | No |
| 8 | Acot8 | 2557 | 0.305 | 0.0288 | No |
| 9 | Isoc1 | 2940 | 0.277 | 0.0179 | No |
| 10 | Msh2 | 3167 | 0.261 | 0.0157 | No |
| 11 | Slc25a19 | 3189 | 0.260 | 0.0259 | No |
| 12 | Gstk1 | 3195 | 0.259 | 0.0369 | No |
| 13 | Pex5 | 3299 | 0.252 | 0.0418 | No |
| 14 | Ywhah | 3362 | 0.249 | 0.0490 | No |
| 15 | Scp2 | 3448 | 0.243 | 0.0545 | No |
| 16 | Smarcc1 | 3529 | 0.238 | 0.0601 | No |
| 17 | Pex6 | 3741 | 0.222 | 0.0571 | No |
| 18 | Ide | 3835 | 0.216 | 0.0610 | No |
| 19 | Cln8 | 4101 | 0.201 | 0.0538 | No |
| 20 | Fdps | 4277 | 0.189 | 0.0516 | No |
| 21 | Hsd3b7 | 4607 | 0.170 | 0.0392 | No |
| 22 | Cdk7 | 4733 | 0.163 | 0.0389 | No |
| 23 | Acsl4 | 4793 | 0.159 | 0.0423 | No |
| 24 | Hmgcl | 5186 | 0.137 | 0.0247 | No |
| 25 | Abcb4 | 5514 | 0.120 | 0.0103 | No |
| 26 | Pex14 | 5881 | 0.103 | -0.0072 | No |
| 27 | Idh1 | 6045 | 0.094 | -0.0129 | No |
| 28 | Cadm1 | 6154 | 0.088 | -0.0156 | No |
| 29 | Vps4b | 6164 | 0.087 | -0.0123 | No |
| 30 | Crat | 6429 | 0.073 | -0.0250 | No |
| 31 | Dlg4 | 6623 | 0.063 | -0.0339 | No |
| 32 | Sema3c | 6692 | 0.060 | -0.0353 | No |
| 33 | Lonp2 | 6725 | 0.057 | -0.0348 | No |
| 34 | Acsl5 | 6930 | 0.047 | -0.0450 | No |
| 35 | Rdh11 | 7619 | 0.015 | -0.0858 | No |
| 36 | Bcl10 | 7671 | 0.012 | -0.0883 | No |
| 37 | Slc25a4 | 8106 | -0.006 | -0.1141 | No |
| 38 | Ephx2 | 8119 | -0.007 | -0.1146 | No |
| 39 | Abcd2 | 8261 | -0.014 | -0.1224 | No |
| 40 | Mlycd | 8493 | -0.024 | -0.1353 | No |
| 41 | Dhrs3 | 8609 | -0.030 | -0.1409 | No |
| 42 | Elovl5 | 9223 | -0.062 | -0.1750 | No |
| 43 | Acox1 | 9365 | -0.071 | -0.1804 | No |
| 44 | Mvp | 9470 | -0.076 | -0.1833 | No |
| 45 | Ercc3 | 9726 | -0.090 | -0.1947 | No |
| 46 | Itgb1bp1 | 9904 | -0.098 | -0.2011 | No |
| 47 | Eci2 | 10079 | -0.106 | -0.2069 | No |
| 48 | Prdx5 | 10116 | -0.107 | -0.2044 | No |
| 49 | Abcb9 | 10127 | -0.108 | -0.2002 | No |
| 50 | Sod2 | 10357 | -0.121 | -0.2087 | No |
| 51 | Gnpat | 10624 | -0.135 | -0.2188 | No |
| 52 | Abcd1 | 11042 | -0.155 | -0.2371 | No |
| 53 | Abcc5 | 11095 | -0.158 | -0.2333 | No |
| 54 | Slc35b2 | 11232 | -0.166 | -0.2342 | No |
| 55 | Cnbp | 11239 | -0.167 | -0.2273 | No |
| 56 | Tspo | 11906 | -0.205 | -0.2584 | No |
| 57 | Pabpc1 | 12230 | -0.222 | -0.2681 | Yes |
| 58 | Atxn1 | 12289 | -0.226 | -0.2617 | Yes |
| 59 | Abcd3 | 12513 | -0.241 | -0.2646 | Yes |
| 60 | Sult2b1 | 12667 | -0.247 | -0.2629 | Yes |
| 61 | Hras | 12766 | -0.254 | -0.2577 | Yes |
| 62 | Hsd17b4 | 12772 | -0.254 | -0.2469 | Yes |
| 63 | Prdx1 | 12901 | -0.260 | -0.2432 | Yes |
| 64 | Acsl1 | 13218 | -0.282 | -0.2498 | Yes |
| 65 | Aldh1a1 | 13322 | -0.290 | -0.2434 | Yes |
| 66 | Aldh9a1 | 13488 | -0.302 | -0.2401 | Yes |
| 67 | Abcb1a | 13692 | -0.315 | -0.2385 | Yes |
| 68 | Cln6 | 14068 | -0.341 | -0.2461 | Yes |
| 69 | Hsd17b11 | 14094 | -0.343 | -0.2326 | Yes |
| 70 | Ercc1 | 14191 | -0.351 | -0.2230 | Yes |
| 71 | Slc25a17 | 14496 | -0.381 | -0.2245 | Yes |
| 72 | Nudt19 | 14776 | -0.407 | -0.2235 | Yes |
| 73 | Pex11a | 14855 | -0.413 | -0.2101 | Yes |
| 74 | Acaa1a | 14999 | -0.430 | -0.1999 | Yes |
| 75 | Cat | 15032 | -0.434 | -0.1828 | Yes |
| 76 | Idh2 | 15226 | -0.460 | -0.1743 | Yes |
| 77 | Siah1a | 15242 | -0.462 | -0.1549 | Yes |
| 78 | Cacna1b | 15313 | -0.471 | -0.1385 | Yes |
| 79 | Ech1 | 15487 | -0.494 | -0.1272 | Yes |
| 80 | Fis1 | 15772 | -0.544 | -0.1205 | Yes |
| 81 | Pex2 | 15893 | -0.563 | -0.1030 | Yes |
| 82 | Pex13 | 16384 | -0.709 | -0.1015 | Yes |
| 83 | Pex11b | 16484 | -0.769 | -0.0737 | Yes |
| 84 | Sod1 | 16651 | -0.977 | -0.0409 | Yes |
| 85 | Ehhadh | 16657 | -0.997 | 0.0025 | Yes |