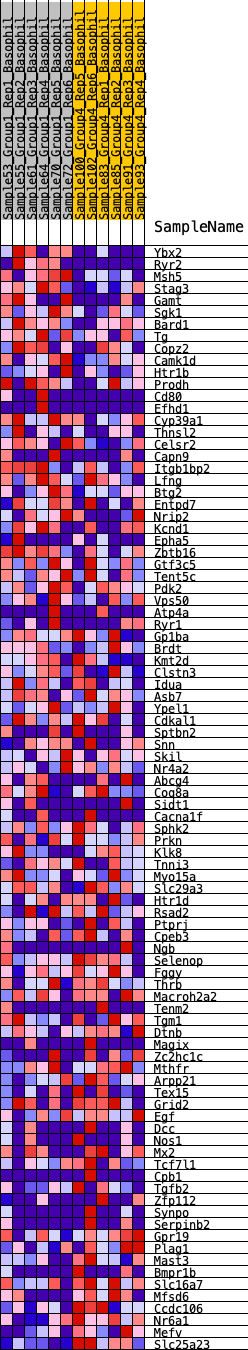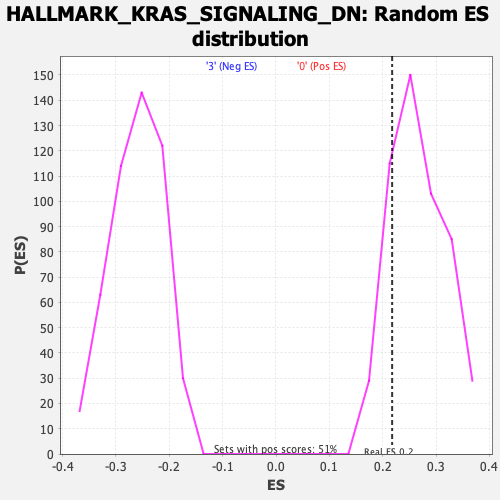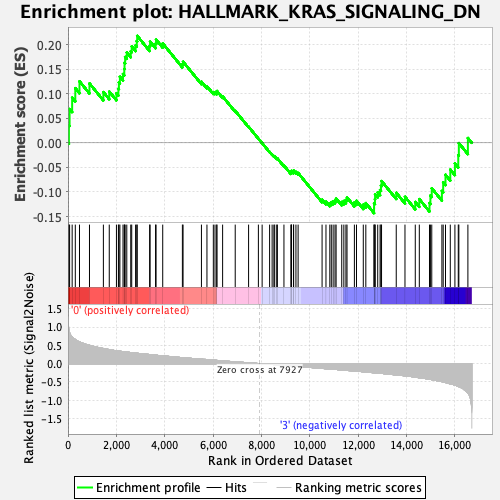
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.21768795 |
| Normalized Enrichment Score (NES) | 0.82124734 |
| Nominal p-value | 0.8003914 |
| FDR q-value | 0.86851096 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ybx2 | 38 | 0.925 | 0.0356 | Yes |
| 2 | Ryr2 | 66 | 0.849 | 0.0688 | Yes |
| 3 | Msh5 | 172 | 0.723 | 0.0921 | Yes |
| 4 | Stag3 | 302 | 0.655 | 0.1112 | Yes |
| 5 | Gamt | 475 | 0.594 | 0.1252 | Yes |
| 6 | Sgk1 | 888 | 0.500 | 0.1209 | Yes |
| 7 | Bard1 | 1463 | 0.413 | 0.1033 | Yes |
| 8 | Tg | 1706 | 0.382 | 0.1043 | Yes |
| 9 | Copz2 | 2003 | 0.351 | 0.1009 | Yes |
| 10 | Camk1d | 2084 | 0.344 | 0.1102 | Yes |
| 11 | Htr1b | 2105 | 0.342 | 0.1230 | Yes |
| 12 | Prodh | 2146 | 0.338 | 0.1344 | Yes |
| 13 | Cd80 | 2275 | 0.328 | 0.1402 | Yes |
| 14 | Efhd1 | 2328 | 0.325 | 0.1504 | Yes |
| 15 | Cyp39a1 | 2338 | 0.324 | 0.1631 | Yes |
| 16 | Thnsl2 | 2363 | 0.321 | 0.1748 | Yes |
| 17 | Celsr2 | 2431 | 0.316 | 0.1837 | Yes |
| 18 | Capn9 | 2595 | 0.302 | 0.1863 | Yes |
| 19 | Itgb1bp2 | 2635 | 0.299 | 0.1962 | Yes |
| 20 | Lfng | 2795 | 0.286 | 0.1983 | Yes |
| 21 | Btg2 | 2844 | 0.285 | 0.2071 | Yes |
| 22 | Entpd7 | 2862 | 0.283 | 0.2177 | Yes |
| 23 | Nrip2 | 3376 | 0.248 | 0.1970 | No |
| 24 | Kcnd1 | 3394 | 0.247 | 0.2061 | No |
| 25 | Epha5 | 3621 | 0.231 | 0.2019 | No |
| 26 | Zbtb16 | 3641 | 0.229 | 0.2102 | No |
| 27 | Gtf3c5 | 3917 | 0.211 | 0.2023 | No |
| 28 | Tent5c | 4730 | 0.163 | 0.1601 | No |
| 29 | Pdk2 | 4758 | 0.162 | 0.1651 | No |
| 30 | Vps50 | 5518 | 0.119 | 0.1243 | No |
| 31 | Atp4a | 5746 | 0.111 | 0.1152 | No |
| 32 | Ryr1 | 6010 | 0.096 | 0.1032 | No |
| 33 | Gp1ba | 6079 | 0.092 | 0.1029 | No |
| 34 | Brdt | 6152 | 0.088 | 0.1022 | No |
| 35 | Kmt2d | 6158 | 0.087 | 0.1055 | No |
| 36 | Clstn3 | 6394 | 0.075 | 0.0944 | No |
| 37 | Idua | 6915 | 0.048 | 0.0651 | No |
| 38 | Asb7 | 7468 | 0.022 | 0.0327 | No |
| 39 | Ypel1 | 7873 | 0.003 | 0.0085 | No |
| 40 | Cdkal1 | 8028 | -0.003 | -0.0006 | No |
| 41 | Sptbn2 | 8337 | -0.018 | -0.0184 | No |
| 42 | Snn | 8441 | -0.022 | -0.0238 | No |
| 43 | Skil | 8514 | -0.025 | -0.0271 | No |
| 44 | Nr4a2 | 8548 | -0.027 | -0.0280 | No |
| 45 | Abcg4 | 8632 | -0.032 | -0.0317 | No |
| 46 | Coq8a | 8658 | -0.033 | -0.0318 | No |
| 47 | Sidt1 | 8930 | -0.047 | -0.0462 | No |
| 48 | Cacna1f | 9217 | -0.062 | -0.0608 | No |
| 49 | Sphk2 | 9238 | -0.063 | -0.0595 | No |
| 50 | Prkn | 9244 | -0.064 | -0.0571 | No |
| 51 | Klk8 | 9328 | -0.068 | -0.0593 | No |
| 52 | Tnni3 | 9329 | -0.068 | -0.0565 | No |
| 53 | Myo15a | 9426 | -0.075 | -0.0593 | No |
| 54 | Slc29a3 | 9516 | -0.078 | -0.0614 | No |
| 55 | Htr1d | 10507 | -0.129 | -0.1157 | No |
| 56 | Rsad2 | 10668 | -0.138 | -0.1197 | No |
| 57 | Ptprj | 10827 | -0.145 | -0.1233 | No |
| 58 | Cpeb3 | 10888 | -0.149 | -0.1207 | No |
| 59 | Ngb | 10964 | -0.154 | -0.1190 | No |
| 60 | Selenop | 11041 | -0.155 | -0.1172 | No |
| 61 | Fggy | 11087 | -0.158 | -0.1134 | No |
| 62 | Thrb | 11320 | -0.171 | -0.1204 | No |
| 63 | Macroh2a2 | 11407 | -0.176 | -0.1184 | No |
| 64 | Tenm2 | 11493 | -0.181 | -0.1161 | No |
| 65 | Tgm1 | 11538 | -0.184 | -0.1112 | No |
| 66 | Dtnb | 11843 | -0.200 | -0.1213 | No |
| 67 | Magix | 11930 | -0.206 | -0.1180 | No |
| 68 | Zc2hc1c | 12211 | -0.222 | -0.1258 | No |
| 69 | Mthfr | 12316 | -0.228 | -0.1227 | No |
| 70 | Arpp21 | 12657 | -0.247 | -0.1331 | No |
| 71 | Tex15 | 12658 | -0.247 | -0.1229 | No |
| 72 | Grid2 | 12693 | -0.249 | -0.1148 | No |
| 73 | Egf | 12700 | -0.250 | -0.1049 | No |
| 74 | Dcc | 12808 | -0.256 | -0.1009 | No |
| 75 | Nos1 | 12906 | -0.261 | -0.0960 | No |
| 76 | Mx2 | 12942 | -0.263 | -0.0874 | No |
| 77 | Tcf7l1 | 12966 | -0.265 | -0.0779 | No |
| 78 | Cpb1 | 13574 | -0.308 | -0.1018 | No |
| 79 | Tgfb2 | 13935 | -0.332 | -0.1099 | No |
| 80 | Zfp112 | 14361 | -0.367 | -0.1204 | No |
| 81 | Synpo | 14528 | -0.385 | -0.1146 | No |
| 82 | Serpinb2 | 14949 | -0.424 | -0.1225 | No |
| 83 | Gpr19 | 14987 | -0.429 | -0.1072 | No |
| 84 | Plag1 | 15043 | -0.436 | -0.0926 | No |
| 85 | Mast3 | 15462 | -0.490 | -0.0977 | No |
| 86 | Bmpr1b | 15515 | -0.499 | -0.0804 | No |
| 87 | Slc16a7 | 15611 | -0.514 | -0.0650 | No |
| 88 | Mfsd6 | 15809 | -0.550 | -0.0543 | No |
| 89 | Ccdc106 | 16000 | -0.583 | -0.0419 | No |
| 90 | Nr6a1 | 16139 | -0.617 | -0.0249 | No |
| 91 | Mefv | 16163 | -0.626 | -0.0006 | No |
| 92 | Slc25a23 | 16537 | -0.800 | 0.0098 | No |