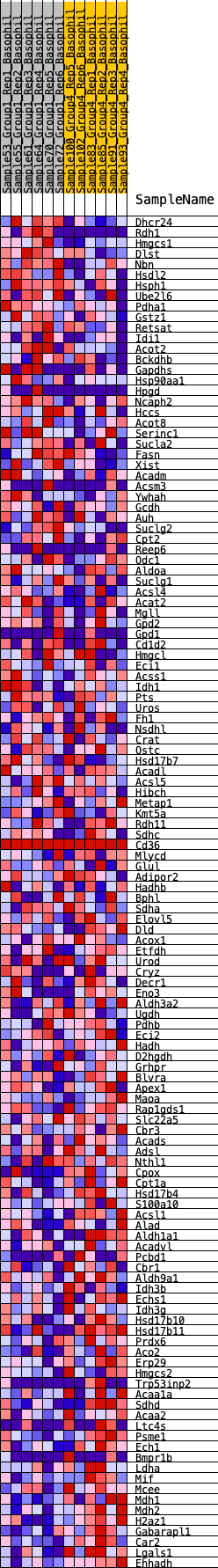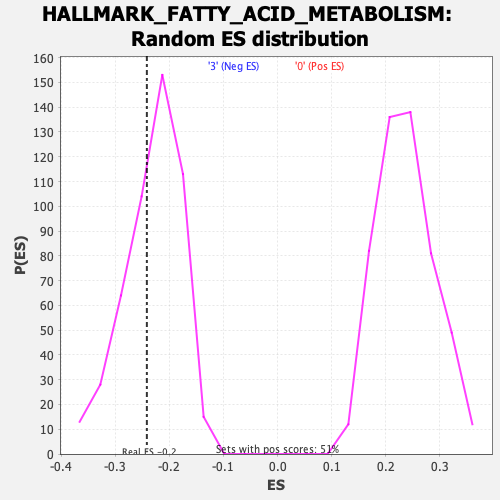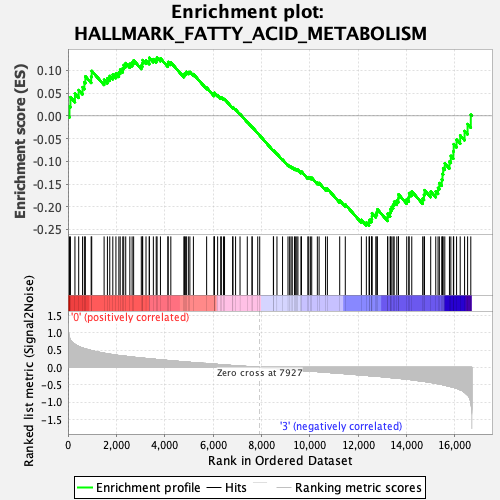
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2414632 |
| Normalized Enrichment Score (NES) | -1.0501431 |
| Nominal p-value | 0.36734694 |
| FDR q-value | 0.61522806 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 61 | 0.862 | 0.0209 | No |
| 2 | Rdh1 | 101 | 0.789 | 0.0411 | No |
| 3 | Hmgcs1 | 287 | 0.664 | 0.0489 | No |
| 4 | Dlst | 442 | 0.601 | 0.0568 | No |
| 5 | Nbn | 602 | 0.559 | 0.0632 | No |
| 6 | Hsdl2 | 677 | 0.541 | 0.0741 | No |
| 7 | Hsph1 | 718 | 0.532 | 0.0869 | No |
| 8 | Ube2l6 | 958 | 0.487 | 0.0864 | No |
| 9 | Pdha1 | 981 | 0.483 | 0.0988 | No |
| 10 | Gstz1 | 1493 | 0.410 | 0.0797 | No |
| 11 | Retsat | 1630 | 0.392 | 0.0827 | No |
| 12 | Idi1 | 1727 | 0.380 | 0.0878 | No |
| 13 | Acot2 | 1853 | 0.366 | 0.0907 | No |
| 14 | Bckdhb | 1982 | 0.353 | 0.0930 | No |
| 15 | Gapdhs | 2104 | 0.342 | 0.0955 | No |
| 16 | Hsp90aa1 | 2158 | 0.338 | 0.1019 | No |
| 17 | Hpgd | 2265 | 0.328 | 0.1049 | No |
| 18 | Ncaph2 | 2305 | 0.326 | 0.1119 | No |
| 19 | Hccs | 2387 | 0.319 | 0.1161 | No |
| 20 | Acot8 | 2557 | 0.305 | 0.1146 | No |
| 21 | Serinc1 | 2651 | 0.298 | 0.1175 | No |
| 22 | Sucla2 | 2714 | 0.293 | 0.1221 | No |
| 23 | Fasn | 3031 | 0.271 | 0.1108 | No |
| 24 | Xist | 3077 | 0.269 | 0.1158 | No |
| 25 | Acadm | 3089 | 0.268 | 0.1227 | No |
| 26 | Acsm3 | 3226 | 0.257 | 0.1219 | No |
| 27 | Ywhah | 3362 | 0.249 | 0.1208 | No |
| 28 | Gcdh | 3363 | 0.249 | 0.1280 | No |
| 29 | Auh | 3530 | 0.238 | 0.1247 | No |
| 30 | Suclg2 | 3648 | 0.229 | 0.1242 | No |
| 31 | Cpt2 | 3683 | 0.227 | 0.1286 | No |
| 32 | Reep6 | 3823 | 0.217 | 0.1264 | No |
| 33 | Odc1 | 4130 | 0.199 | 0.1136 | No |
| 34 | Aldoa | 4142 | 0.198 | 0.1186 | No |
| 35 | Suclg1 | 4252 | 0.190 | 0.1175 | No |
| 36 | Acsl4 | 4793 | 0.159 | 0.0895 | No |
| 37 | Acat2 | 4815 | 0.158 | 0.0927 | No |
| 38 | Mgll | 4865 | 0.156 | 0.0942 | No |
| 39 | Gpd2 | 4895 | 0.154 | 0.0969 | No |
| 40 | Gpd1 | 4976 | 0.150 | 0.0963 | No |
| 41 | Cd1d2 | 5040 | 0.146 | 0.0967 | No |
| 42 | Hmgcl | 5186 | 0.137 | 0.0919 | No |
| 43 | Eci1 | 5733 | 0.111 | 0.0621 | No |
| 44 | Acss1 | 6041 | 0.094 | 0.0462 | No |
| 45 | Idh1 | 6045 | 0.094 | 0.0487 | No |
| 46 | Pts | 6056 | 0.093 | 0.0508 | No |
| 47 | Uros | 6188 | 0.086 | 0.0454 | No |
| 48 | Fh1 | 6316 | 0.079 | 0.0399 | No |
| 49 | Nsdhl | 6340 | 0.077 | 0.0408 | No |
| 50 | Crat | 6429 | 0.073 | 0.0375 | No |
| 51 | Ostc | 6473 | 0.071 | 0.0370 | No |
| 52 | Hsd17b7 | 6808 | 0.054 | 0.0184 | No |
| 53 | Acadl | 6822 | 0.053 | 0.0191 | No |
| 54 | Acsl5 | 6930 | 0.047 | 0.0140 | No |
| 55 | Hibch | 7114 | 0.038 | 0.0040 | No |
| 56 | Metap1 | 7414 | 0.026 | -0.0133 | No |
| 57 | Kmt5a | 7608 | 0.015 | -0.0245 | No |
| 58 | Rdh11 | 7619 | 0.015 | -0.0247 | No |
| 59 | Sdhc | 7846 | 0.004 | -0.0382 | No |
| 60 | Cd36 | 7928 | 0.000 | -0.0431 | No |
| 61 | Mlycd | 8493 | -0.024 | -0.0764 | No |
| 62 | Glul | 8500 | -0.024 | -0.0761 | No |
| 63 | Adipor2 | 8641 | -0.032 | -0.0836 | No |
| 64 | Hadhb | 8872 | -0.044 | -0.0963 | No |
| 65 | Bphl | 9098 | -0.056 | -0.1083 | No |
| 66 | Sdha | 9166 | -0.059 | -0.1106 | No |
| 67 | Elovl5 | 9223 | -0.062 | -0.1122 | No |
| 68 | Dld | 9273 | -0.065 | -0.1133 | No |
| 69 | Acox1 | 9365 | -0.071 | -0.1168 | No |
| 70 | Etfdh | 9401 | -0.073 | -0.1168 | No |
| 71 | Urod | 9460 | -0.076 | -0.1181 | No |
| 72 | Cryz | 9508 | -0.078 | -0.1187 | No |
| 73 | Decr1 | 9633 | -0.085 | -0.1238 | No |
| 74 | Eno3 | 9655 | -0.086 | -0.1226 | No |
| 75 | Aldh3a2 | 9918 | -0.098 | -0.1356 | No |
| 76 | Ugdh | 9955 | -0.100 | -0.1349 | No |
| 77 | Pdhb | 10023 | -0.103 | -0.1360 | No |
| 78 | Eci2 | 10079 | -0.106 | -0.1363 | No |
| 79 | Hadh | 10311 | -0.118 | -0.1469 | No |
| 80 | D2hgdh | 10384 | -0.122 | -0.1477 | No |
| 81 | Grhpr | 10651 | -0.137 | -0.1599 | No |
| 82 | Blvra | 10726 | -0.140 | -0.1603 | No |
| 83 | Apex1 | 11236 | -0.166 | -0.1863 | No |
| 84 | Maoa | 11467 | -0.179 | -0.1950 | No |
| 85 | Rap1gds1 | 12132 | -0.216 | -0.2289 | No |
| 86 | Slc22a5 | 12338 | -0.229 | -0.2348 | Yes |
| 87 | Cbr3 | 12450 | -0.237 | -0.2347 | Yes |
| 88 | Acads | 12469 | -0.238 | -0.2290 | Yes |
| 89 | Adsl | 12550 | -0.242 | -0.2269 | Yes |
| 90 | Nthl1 | 12568 | -0.243 | -0.2210 | Yes |
| 91 | Cpox | 12571 | -0.243 | -0.2142 | Yes |
| 92 | Cpt1a | 12729 | -0.251 | -0.2165 | Yes |
| 93 | Hsd17b4 | 12772 | -0.254 | -0.2118 | Yes |
| 94 | S100a10 | 12793 | -0.256 | -0.2057 | Yes |
| 95 | Acsl1 | 13218 | -0.282 | -0.2232 | Yes |
| 96 | Alad | 13222 | -0.282 | -0.2153 | Yes |
| 97 | Aldh1a1 | 13322 | -0.290 | -0.2130 | Yes |
| 98 | Acadvl | 13335 | -0.291 | -0.2054 | Yes |
| 99 | Pcbd1 | 13390 | -0.295 | -0.2003 | Yes |
| 100 | Cbr1 | 13447 | -0.298 | -0.1951 | Yes |
| 101 | Aldh9a1 | 13488 | -0.302 | -0.1889 | Yes |
| 102 | Idh3b | 13597 | -0.310 | -0.1866 | Yes |
| 103 | Echs1 | 13663 | -0.313 | -0.1816 | Yes |
| 104 | Idh3g | 13666 | -0.314 | -0.1727 | Yes |
| 105 | Hsd17b10 | 14007 | -0.338 | -0.1836 | Yes |
| 106 | Hsd17b11 | 14094 | -0.343 | -0.1790 | Yes |
| 107 | Prdx6 | 14106 | -0.344 | -0.1698 | Yes |
| 108 | Aco2 | 14216 | -0.353 | -0.1663 | Yes |
| 109 | Erp29 | 14667 | -0.398 | -0.1821 | Yes |
| 110 | Hmgcs2 | 14721 | -0.402 | -0.1738 | Yes |
| 111 | Trp53inp2 | 14743 | -0.404 | -0.1636 | Yes |
| 112 | Acaa1a | 14999 | -0.430 | -0.1667 | Yes |
| 113 | Sdhd | 15208 | -0.457 | -0.1662 | Yes |
| 114 | Acaa2 | 15303 | -0.470 | -0.1584 | Yes |
| 115 | Ltc4s | 15356 | -0.478 | -0.1479 | Yes |
| 116 | Psme1 | 15456 | -0.488 | -0.1399 | Yes |
| 117 | Ech1 | 15487 | -0.494 | -0.1276 | Yes |
| 118 | Bmpr1b | 15515 | -0.499 | -0.1150 | Yes |
| 119 | Ldha | 15582 | -0.510 | -0.1044 | Yes |
| 120 | Mif | 15774 | -0.545 | -0.1004 | Yes |
| 121 | Mcee | 15832 | -0.553 | -0.0880 | Yes |
| 122 | Mdh1 | 15937 | -0.570 | -0.0780 | Yes |
| 123 | Mdh2 | 15950 | -0.573 | -0.0624 | Yes |
| 124 | H2az1 | 16069 | -0.599 | -0.0524 | Yes |
| 125 | Gabarapl1 | 16219 | -0.643 | -0.0430 | Yes |
| 126 | Car2 | 16400 | -0.719 | -0.0333 | Yes |
| 127 | Lgals1 | 16525 | -0.796 | -0.0181 | Yes |
| 128 | Ehhadh | 16657 | -0.997 | 0.0025 | Yes |