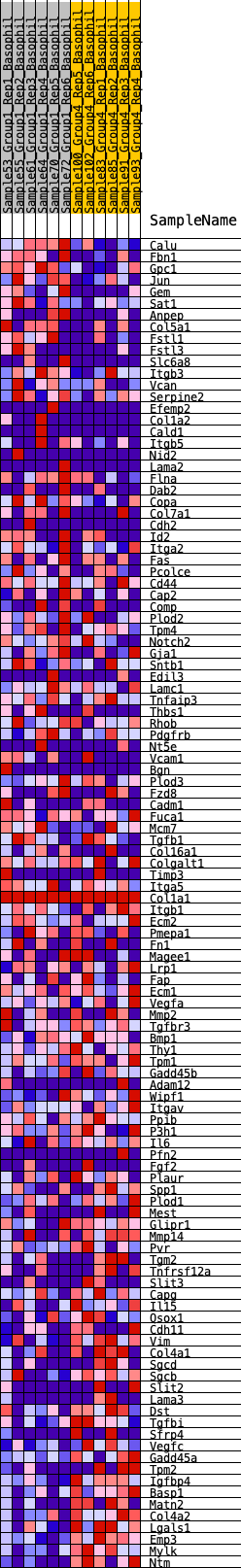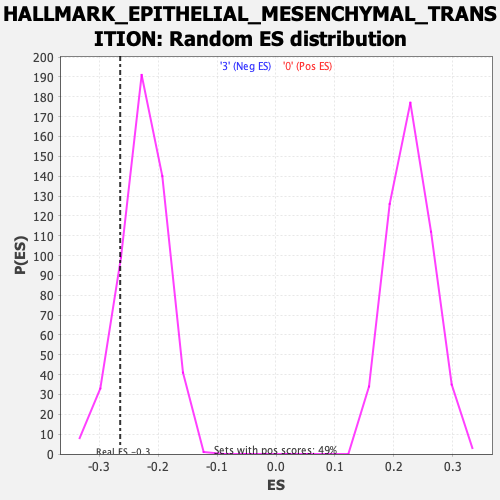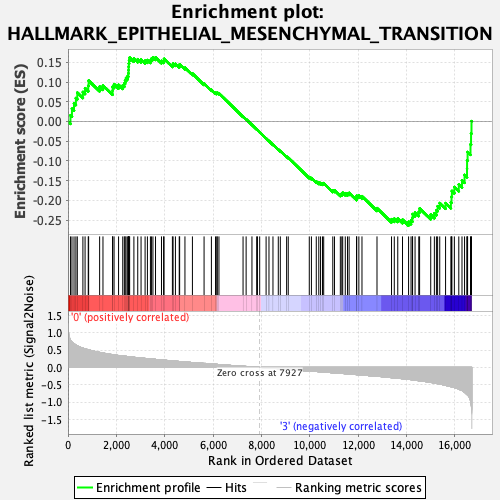
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.2641297 |
| Normalized Enrichment Score (NES) | -1.1725776 |
| Nominal p-value | 0.15594542 |
| FDR q-value | 0.47775227 |
| FWER p-Value | 0.969 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Calu | 102 | 0.787 | 0.0157 | No |
| 2 | Fbn1 | 164 | 0.728 | 0.0323 | No |
| 3 | Gpc1 | 249 | 0.679 | 0.0461 | No |
| 4 | Jun | 327 | 0.644 | 0.0594 | No |
| 5 | Gem | 385 | 0.621 | 0.0732 | No |
| 6 | Sat1 | 616 | 0.556 | 0.0748 | No |
| 7 | Anpep | 706 | 0.533 | 0.0843 | No |
| 8 | Col5a1 | 837 | 0.508 | 0.0905 | No |
| 9 | Fstl1 | 853 | 0.505 | 0.1037 | No |
| 10 | Fstl3 | 1308 | 0.433 | 0.0883 | No |
| 11 | Slc6a8 | 1446 | 0.414 | 0.0916 | No |
| 12 | Itgb3 | 1843 | 0.367 | 0.0779 | No |
| 13 | Vcan | 1846 | 0.367 | 0.0880 | No |
| 14 | Serpine2 | 1905 | 0.359 | 0.0945 | No |
| 15 | Efemp2 | 2091 | 0.343 | 0.0929 | No |
| 16 | Col1a2 | 2264 | 0.328 | 0.0916 | No |
| 17 | Cald1 | 2329 | 0.325 | 0.0968 | No |
| 18 | Itgb5 | 2361 | 0.322 | 0.1039 | No |
| 19 | Nid2 | 2401 | 0.318 | 0.1104 | No |
| 20 | Lama2 | 2467 | 0.313 | 0.1151 | No |
| 21 | Flna | 2496 | 0.310 | 0.1221 | No |
| 22 | Dab2 | 2504 | 0.309 | 0.1303 | No |
| 23 | Copa | 2509 | 0.309 | 0.1386 | No |
| 24 | Col7a1 | 2515 | 0.309 | 0.1469 | No |
| 25 | Cdh2 | 2527 | 0.308 | 0.1548 | No |
| 26 | Id2 | 2549 | 0.306 | 0.1620 | No |
| 27 | Itga2 | 2725 | 0.292 | 0.1596 | No |
| 28 | Fas | 2884 | 0.281 | 0.1578 | No |
| 29 | Pcolce | 3021 | 0.271 | 0.1572 | No |
| 30 | Cd44 | 3188 | 0.260 | 0.1544 | No |
| 31 | Cap2 | 3284 | 0.254 | 0.1557 | No |
| 32 | Comp | 3408 | 0.246 | 0.1552 | No |
| 33 | Plod2 | 3450 | 0.243 | 0.1595 | No |
| 34 | Tpm4 | 3512 | 0.239 | 0.1624 | No |
| 35 | Notch2 | 3619 | 0.231 | 0.1624 | No |
| 36 | Gja1 | 3866 | 0.214 | 0.1536 | No |
| 37 | Sntb1 | 3953 | 0.209 | 0.1542 | No |
| 38 | Edil3 | 3972 | 0.208 | 0.1589 | No |
| 39 | Lamc1 | 4317 | 0.187 | 0.1433 | No |
| 40 | Tnfaip3 | 4345 | 0.185 | 0.1469 | No |
| 41 | Thbs1 | 4442 | 0.179 | 0.1460 | No |
| 42 | Rhob | 4600 | 0.170 | 0.1413 | No |
| 43 | Pdgfrb | 4617 | 0.169 | 0.1450 | No |
| 44 | Nt5e | 4836 | 0.157 | 0.1363 | No |
| 45 | Vcam1 | 5145 | 0.140 | 0.1216 | No |
| 46 | Bgn | 5627 | 0.115 | 0.0958 | No |
| 47 | Plod3 | 5930 | 0.100 | 0.0804 | No |
| 48 | Fzd8 | 6101 | 0.091 | 0.0726 | No |
| 49 | Cadm1 | 6154 | 0.088 | 0.0719 | No |
| 50 | Fuca1 | 6172 | 0.087 | 0.0733 | No |
| 51 | Mcm7 | 6242 | 0.083 | 0.0715 | No |
| 52 | Tgfb1 | 7239 | 0.034 | 0.0124 | No |
| 53 | Col16a1 | 7366 | 0.028 | 0.0055 | No |
| 54 | Colgalt1 | 7607 | 0.015 | -0.0085 | No |
| 55 | Timp3 | 7803 | 0.006 | -0.0201 | No |
| 56 | Itga5 | 7834 | 0.004 | -0.0218 | No |
| 57 | Col1a1 | 7927 | 0.000 | -0.0273 | No |
| 58 | Itgb1 | 8192 | -0.011 | -0.0429 | No |
| 59 | Ecm2 | 8311 | -0.017 | -0.0496 | No |
| 60 | Pmepa1 | 8476 | -0.023 | -0.0588 | No |
| 61 | Fn1 | 8696 | -0.035 | -0.0711 | No |
| 62 | Magee1 | 8780 | -0.039 | -0.0750 | No |
| 63 | Lrp1 | 9041 | -0.053 | -0.0892 | No |
| 64 | Fap | 9108 | -0.056 | -0.0916 | No |
| 65 | Ecm1 | 9978 | -0.101 | -0.1412 | No |
| 66 | Vegfa | 10066 | -0.105 | -0.1435 | No |
| 67 | Mmp2 | 10267 | -0.116 | -0.1524 | No |
| 68 | Tgfbr3 | 10371 | -0.121 | -0.1552 | No |
| 69 | Bmp1 | 10444 | -0.125 | -0.1560 | No |
| 70 | Thy1 | 10524 | -0.130 | -0.1572 | No |
| 71 | Tpm1 | 10574 | -0.133 | -0.1565 | No |
| 72 | Gadd45b | 10950 | -0.153 | -0.1748 | No |
| 73 | Adam12 | 11019 | -0.154 | -0.1746 | No |
| 74 | Wipf1 | 11262 | -0.168 | -0.1846 | No |
| 75 | Itgav | 11323 | -0.171 | -0.1834 | No |
| 76 | Ppib | 11360 | -0.174 | -0.1808 | No |
| 77 | P3h1 | 11466 | -0.179 | -0.1821 | No |
| 78 | Il6 | 11559 | -0.185 | -0.1825 | No |
| 79 | Pfn2 | 11625 | -0.189 | -0.1812 | No |
| 80 | Fgf2 | 11931 | -0.206 | -0.1938 | No |
| 81 | Plaur | 11940 | -0.206 | -0.1886 | No |
| 82 | Spp1 | 12028 | -0.211 | -0.1880 | No |
| 83 | Plod1 | 12157 | -0.217 | -0.1896 | No |
| 84 | Mest | 12778 | -0.255 | -0.2199 | No |
| 85 | Glipr1 | 13379 | -0.294 | -0.2479 | No |
| 86 | Mmp14 | 13493 | -0.303 | -0.2463 | No |
| 87 | Pvr | 13637 | -0.312 | -0.2463 | No |
| 88 | Tgm2 | 13833 | -0.324 | -0.2491 | No |
| 89 | Tnfrsf12a | 14084 | -0.343 | -0.2546 | Yes |
| 90 | Slit3 | 14186 | -0.351 | -0.2509 | Yes |
| 91 | Capg | 14244 | -0.355 | -0.2445 | Yes |
| 92 | Il15 | 14250 | -0.356 | -0.2349 | Yes |
| 93 | Qsox1 | 14352 | -0.366 | -0.2308 | Yes |
| 94 | Cdh11 | 14506 | -0.382 | -0.2294 | Yes |
| 95 | Vim | 14540 | -0.386 | -0.2207 | Yes |
| 96 | Col4a1 | 15000 | -0.430 | -0.2364 | Yes |
| 97 | Sgcd | 15150 | -0.451 | -0.2329 | Yes |
| 98 | Sgcb | 15235 | -0.461 | -0.2251 | Yes |
| 99 | Slit2 | 15285 | -0.467 | -0.2151 | Yes |
| 100 | Lama3 | 15373 | -0.481 | -0.2069 | Yes |
| 101 | Dst | 15614 | -0.515 | -0.2071 | Yes |
| 102 | Tgfbi | 15834 | -0.554 | -0.2049 | Yes |
| 103 | Sfrp4 | 15860 | -0.557 | -0.1909 | Yes |
| 104 | Vegfc | 15875 | -0.559 | -0.1762 | Yes |
| 105 | Gadd45a | 15977 | -0.578 | -0.1662 | Yes |
| 106 | Tpm2 | 16161 | -0.626 | -0.1599 | Yes |
| 107 | Igfbp4 | 16293 | -0.664 | -0.1493 | Yes |
| 108 | Basp1 | 16399 | -0.719 | -0.1356 | Yes |
| 109 | Matn2 | 16501 | -0.780 | -0.1200 | Yes |
| 110 | Col4a2 | 16504 | -0.781 | -0.0984 | Yes |
| 111 | Lgals1 | 16525 | -0.796 | -0.0775 | Yes |
| 112 | Emp3 | 16645 | -0.957 | -0.0581 | Yes |
| 113 | Mylk | 16668 | -1.055 | -0.0301 | Yes |
| 114 | Ntm | 16686 | -1.147 | 0.0008 | Yes |