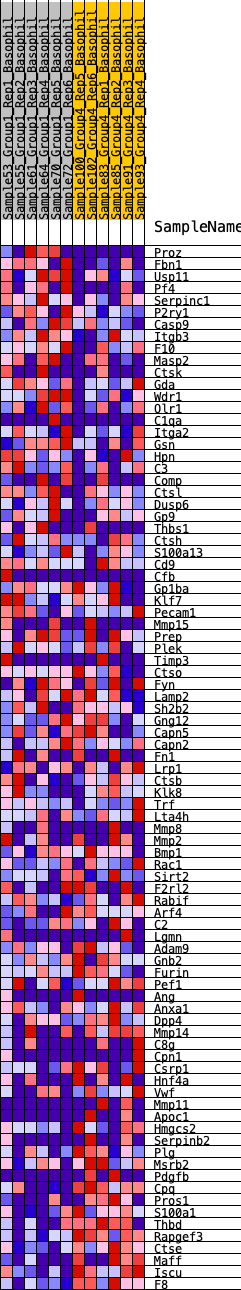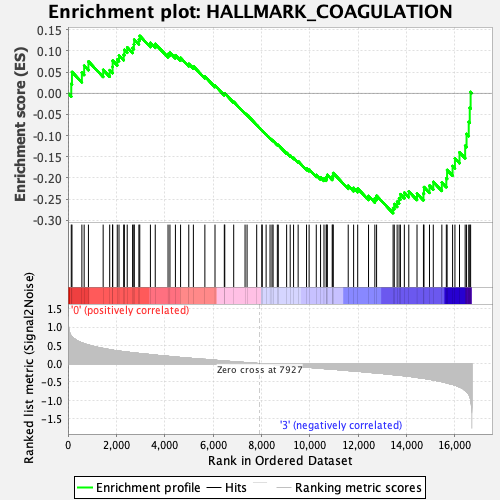
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.2833076 |
| Normalized Enrichment Score (NES) | -1.1983194 |
| Nominal p-value | 0.128 |
| FDR q-value | 0.456384 |
| FWER p-Value | 0.955 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Proz | 135 | 0.749 | 0.0223 | No |
| 2 | Fbn1 | 164 | 0.728 | 0.0502 | No |
| 3 | Usp11 | 574 | 0.565 | 0.0485 | No |
| 4 | Pf4 | 668 | 0.543 | 0.0650 | No |
| 5 | Serpinc1 | 845 | 0.507 | 0.0750 | No |
| 6 | P2ry1 | 1454 | 0.414 | 0.0552 | No |
| 7 | Casp9 | 1724 | 0.380 | 0.0544 | No |
| 8 | Itgb3 | 1843 | 0.367 | 0.0623 | No |
| 9 | F10 | 1848 | 0.367 | 0.0769 | No |
| 10 | Masp2 | 2037 | 0.347 | 0.0797 | No |
| 11 | Ctsk | 2109 | 0.341 | 0.0893 | No |
| 12 | Gda | 2303 | 0.326 | 0.0909 | No |
| 13 | Wdr1 | 2331 | 0.324 | 0.1025 | No |
| 14 | Olr1 | 2449 | 0.315 | 0.1082 | No |
| 15 | C1qa | 2671 | 0.296 | 0.1069 | No |
| 16 | Itga2 | 2725 | 0.292 | 0.1156 | No |
| 17 | Gsn | 2738 | 0.291 | 0.1267 | No |
| 18 | Hpn | 2932 | 0.277 | 0.1264 | No |
| 19 | C3 | 2968 | 0.275 | 0.1354 | No |
| 20 | Comp | 3408 | 0.246 | 0.1190 | No |
| 21 | Ctsl | 3608 | 0.232 | 0.1165 | No |
| 22 | Dusp6 | 4137 | 0.198 | 0.0927 | No |
| 23 | Gp9 | 4216 | 0.193 | 0.0959 | No |
| 24 | Thbs1 | 4442 | 0.179 | 0.0896 | No |
| 25 | Ctsh | 4655 | 0.167 | 0.0836 | No |
| 26 | S100a13 | 4994 | 0.149 | 0.0693 | No |
| 27 | Cd9 | 5185 | 0.138 | 0.0635 | No |
| 28 | Cfb | 5660 | 0.115 | 0.0396 | No |
| 29 | Gp1ba | 6079 | 0.092 | 0.0182 | No |
| 30 | Klf7 | 6464 | 0.071 | -0.0020 | No |
| 31 | Pecam1 | 6483 | 0.070 | -0.0003 | No |
| 32 | Mmp15 | 6852 | 0.051 | -0.0203 | No |
| 33 | Prep | 7322 | 0.030 | -0.0474 | No |
| 34 | Plek | 7405 | 0.026 | -0.0513 | No |
| 35 | Timp3 | 7803 | 0.006 | -0.0749 | No |
| 36 | Ctso | 8014 | -0.002 | -0.0875 | No |
| 37 | Fyn | 8032 | -0.003 | -0.0884 | No |
| 38 | Lamp2 | 8193 | -0.011 | -0.0976 | No |
| 39 | Sh2b2 | 8355 | -0.018 | -0.1065 | No |
| 40 | Gng12 | 8435 | -0.021 | -0.1104 | No |
| 41 | Capn5 | 8481 | -0.023 | -0.1122 | No |
| 42 | Capn2 | 8657 | -0.033 | -0.1214 | No |
| 43 | Fn1 | 8696 | -0.035 | -0.1222 | No |
| 44 | Lrp1 | 9041 | -0.053 | -0.1408 | No |
| 45 | Ctsb | 9189 | -0.061 | -0.1472 | No |
| 46 | Klk8 | 9328 | -0.068 | -0.1527 | No |
| 47 | Trf | 9517 | -0.078 | -0.1608 | No |
| 48 | Lta4h | 9864 | -0.096 | -0.1778 | No |
| 49 | Mmp8 | 9971 | -0.101 | -0.1801 | No |
| 50 | Mmp2 | 10267 | -0.116 | -0.1931 | No |
| 51 | Bmp1 | 10444 | -0.125 | -0.1986 | No |
| 52 | Rac1 | 10586 | -0.133 | -0.2017 | No |
| 53 | Sirt2 | 10675 | -0.138 | -0.2014 | No |
| 54 | F2rl2 | 10702 | -0.139 | -0.1973 | No |
| 55 | Rabif | 10723 | -0.140 | -0.1928 | No |
| 56 | Arf4 | 10926 | -0.152 | -0.1988 | No |
| 57 | C2 | 10955 | -0.153 | -0.1943 | No |
| 58 | Lgmn | 10965 | -0.154 | -0.1886 | No |
| 59 | Adam9 | 11588 | -0.187 | -0.2184 | No |
| 60 | Gnb2 | 11811 | -0.199 | -0.2237 | No |
| 61 | Furin | 11980 | -0.208 | -0.2253 | No |
| 62 | Pef1 | 12428 | -0.235 | -0.2427 | No |
| 63 | Ang | 12689 | -0.248 | -0.2483 | No |
| 64 | Anxa1 | 12761 | -0.254 | -0.2423 | No |
| 65 | Dpp4 | 13444 | -0.298 | -0.2712 | Yes |
| 66 | Mmp14 | 13493 | -0.303 | -0.2618 | Yes |
| 67 | C8g | 13611 | -0.311 | -0.2562 | Yes |
| 68 | Cpn1 | 13691 | -0.315 | -0.2482 | Yes |
| 69 | Csrp1 | 13747 | -0.319 | -0.2385 | Yes |
| 70 | Hnf4a | 13909 | -0.330 | -0.2348 | Yes |
| 71 | Vwf | 14093 | -0.343 | -0.2319 | Yes |
| 72 | Mmp11 | 14433 | -0.375 | -0.2371 | Yes |
| 73 | Apoc1 | 14699 | -0.400 | -0.2368 | Yes |
| 74 | Hmgcs2 | 14721 | -0.402 | -0.2217 | Yes |
| 75 | Serpinb2 | 14949 | -0.424 | -0.2182 | Yes |
| 76 | Plg | 15106 | -0.444 | -0.2095 | Yes |
| 77 | Msrb2 | 15460 | -0.489 | -0.2109 | Yes |
| 78 | Pdgfb | 15645 | -0.521 | -0.2008 | Yes |
| 79 | Cpq | 15677 | -0.527 | -0.1813 | Yes |
| 80 | Pros1 | 15904 | -0.565 | -0.1719 | Yes |
| 81 | S100a1 | 16001 | -0.584 | -0.1540 | Yes |
| 82 | Thbd | 16190 | -0.633 | -0.1396 | Yes |
| 83 | Rapgef3 | 16424 | -0.730 | -0.1240 | Yes |
| 84 | Ctse | 16482 | -0.768 | -0.0962 | Yes |
| 85 | Maff | 16572 | -0.831 | -0.0678 | Yes |
| 86 | Iscu | 16614 | -0.880 | -0.0345 | Yes |
| 87 | F8 | 16650 | -0.975 | 0.0029 | Yes |