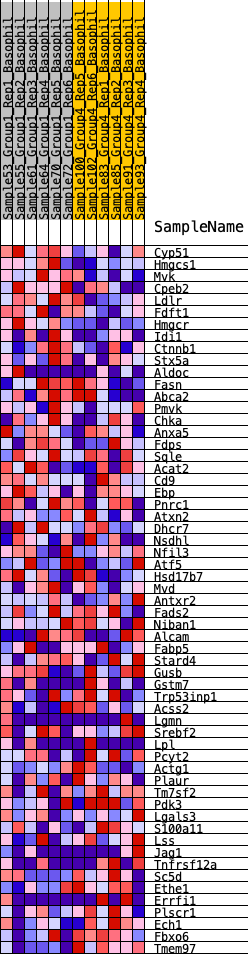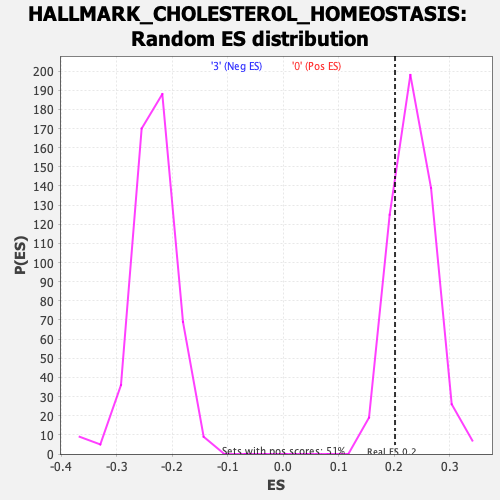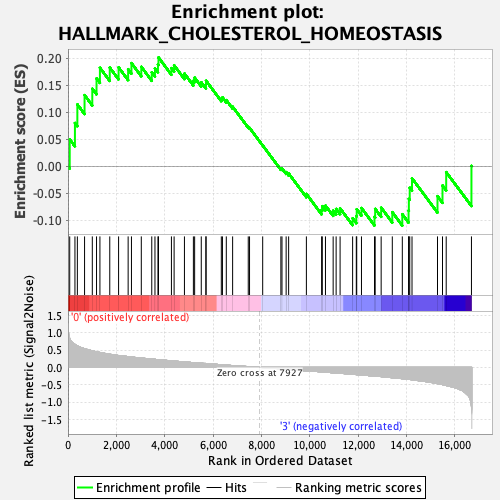
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.20116083 |
| Normalized Enrichment Score (NES) | 0.86299837 |
| Nominal p-value | 0.8249027 |
| FDR q-value | 0.958852 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 71 | 0.833 | 0.0499 | Yes |
| 2 | Hmgcs1 | 287 | 0.664 | 0.0801 | Yes |
| 3 | Mvk | 387 | 0.619 | 0.1144 | Yes |
| 4 | Cpeb2 | 685 | 0.538 | 0.1315 | Yes |
| 5 | Ldlr | 1005 | 0.478 | 0.1434 | Yes |
| 6 | Fdft1 | 1180 | 0.452 | 0.1624 | Yes |
| 7 | Hmgcr | 1320 | 0.432 | 0.1821 | Yes |
| 8 | Idi1 | 1727 | 0.380 | 0.1824 | Yes |
| 9 | Ctnnb1 | 2092 | 0.343 | 0.1828 | Yes |
| 10 | Stx5a | 2487 | 0.311 | 0.1794 | Yes |
| 11 | Aldoc | 2624 | 0.300 | 0.1907 | Yes |
| 12 | Fasn | 3031 | 0.271 | 0.1839 | Yes |
| 13 | Abca2 | 3464 | 0.242 | 0.1736 | Yes |
| 14 | Pmvk | 3596 | 0.233 | 0.1809 | Yes |
| 15 | Chka | 3720 | 0.223 | 0.1880 | Yes |
| 16 | Anxa5 | 3742 | 0.222 | 0.2012 | Yes |
| 17 | Fdps | 4277 | 0.189 | 0.1814 | No |
| 18 | Sqle | 4388 | 0.183 | 0.1867 | No |
| 19 | Acat2 | 4815 | 0.158 | 0.1713 | No |
| 20 | Cd9 | 5185 | 0.138 | 0.1581 | No |
| 21 | Ebp | 5237 | 0.135 | 0.1638 | No |
| 22 | Pnrc1 | 5511 | 0.120 | 0.1552 | No |
| 23 | Atxn2 | 5704 | 0.113 | 0.1510 | No |
| 24 | Dhcr7 | 5708 | 0.113 | 0.1582 | No |
| 25 | Nsdhl | 6340 | 0.077 | 0.1253 | No |
| 26 | Nfil3 | 6387 | 0.075 | 0.1274 | No |
| 27 | Atf5 | 6546 | 0.068 | 0.1223 | No |
| 28 | Hsd17b7 | 6808 | 0.054 | 0.1101 | No |
| 29 | Mvd | 7454 | 0.023 | 0.0729 | No |
| 30 | Antxr2 | 7507 | 0.021 | 0.0711 | No |
| 31 | Fads2 | 8051 | -0.004 | 0.0387 | No |
| 32 | Niban1 | 8803 | -0.040 | -0.0039 | No |
| 33 | Alcam | 8849 | -0.042 | -0.0038 | No |
| 34 | Fabp5 | 9021 | -0.052 | -0.0107 | No |
| 35 | Stard4 | 9124 | -0.057 | -0.0131 | No |
| 36 | Gusb | 9858 | -0.095 | -0.0509 | No |
| 37 | Gstm7 | 10488 | -0.128 | -0.0804 | No |
| 38 | Trp53inp1 | 10516 | -0.130 | -0.0736 | No |
| 39 | Acss2 | 10647 | -0.137 | -0.0725 | No |
| 40 | Lgmn | 10965 | -0.154 | -0.0816 | No |
| 41 | Srebf2 | 11088 | -0.158 | -0.0787 | No |
| 42 | Lpl | 11252 | -0.167 | -0.0776 | No |
| 43 | Pcyt2 | 11770 | -0.196 | -0.0959 | No |
| 44 | Actg1 | 11922 | -0.206 | -0.0917 | No |
| 45 | Plaur | 11940 | -0.206 | -0.0793 | No |
| 46 | Tm7sf2 | 12136 | -0.216 | -0.0770 | No |
| 47 | Pdk3 | 12675 | -0.248 | -0.0932 | No |
| 48 | Lgals3 | 12703 | -0.250 | -0.0786 | No |
| 49 | S100a11 | 12950 | -0.263 | -0.0762 | No |
| 50 | Lss | 13411 | -0.296 | -0.0846 | No |
| 51 | Jag1 | 13825 | -0.323 | -0.0884 | No |
| 52 | Tnfrsf12a | 14084 | -0.343 | -0.0817 | No |
| 53 | Sc5d | 14096 | -0.344 | -0.0600 | No |
| 54 | Ethe1 | 14132 | -0.347 | -0.0396 | No |
| 55 | Errfi1 | 14226 | -0.354 | -0.0221 | No |
| 56 | Plscr1 | 15280 | -0.467 | -0.0551 | No |
| 57 | Ech1 | 15487 | -0.494 | -0.0353 | No |
| 58 | Fbxo6 | 15640 | -0.520 | -0.0107 | No |
| 59 | Tmem97 | 16684 | -1.143 | 0.0009 | No |