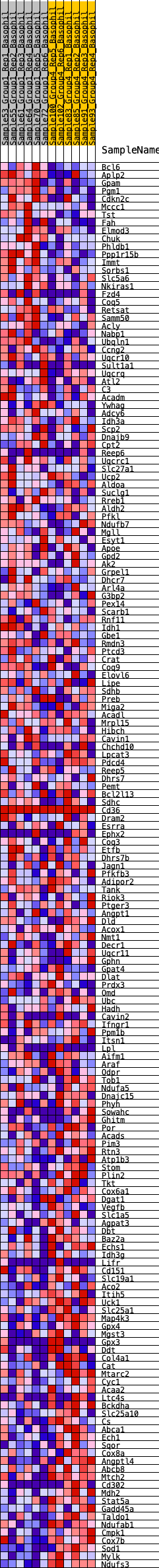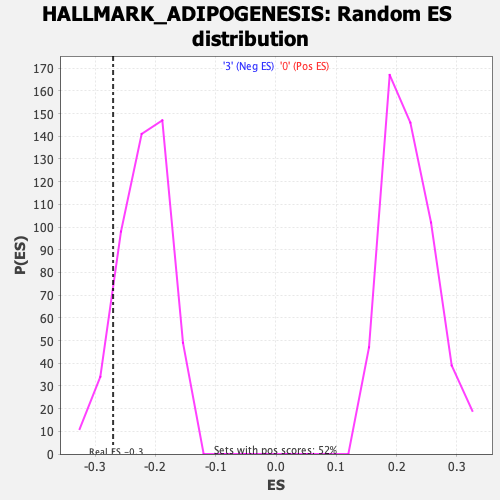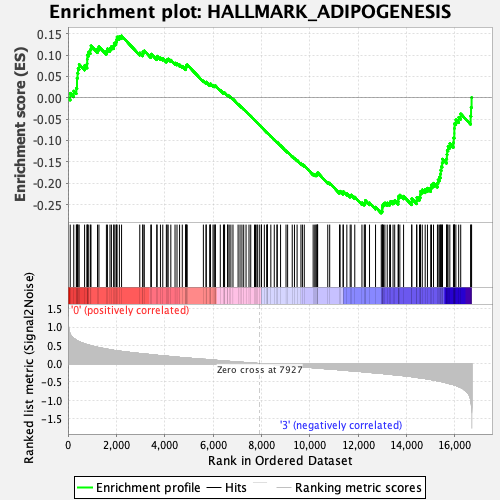
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group4.Basophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ADIPOGENESIS |
| Enrichment Score (ES) | -0.2700746 |
| Normalized Enrichment Score (NES) | -1.2324288 |
| Nominal p-value | 0.11875 |
| FDR q-value | 0.4145673 |
| FWER p-Value | 0.939 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bcl6 | 92 | 0.799 | 0.0107 | No |
| 2 | Aplp2 | 238 | 0.683 | 0.0158 | No |
| 3 | Gpam | 345 | 0.634 | 0.0223 | No |
| 4 | Pgm1 | 372 | 0.626 | 0.0335 | No |
| 5 | Cdkn2c | 373 | 0.626 | 0.0462 | No |
| 6 | Mccc1 | 395 | 0.616 | 0.0574 | No |
| 7 | Tst | 416 | 0.609 | 0.0686 | No |
| 8 | Fah | 461 | 0.597 | 0.0781 | No |
| 9 | Elmod3 | 686 | 0.538 | 0.0755 | No |
| 10 | Chuk | 790 | 0.519 | 0.0798 | No |
| 11 | Phldb1 | 795 | 0.518 | 0.0901 | No |
| 12 | Ppp1r15b | 800 | 0.516 | 0.1003 | No |
| 13 | Immt | 856 | 0.505 | 0.1073 | No |
| 14 | Sorbs1 | 927 | 0.493 | 0.1131 | No |
| 15 | Slc5a6 | 950 | 0.489 | 0.1217 | No |
| 16 | Nkiras1 | 1221 | 0.444 | 0.1144 | No |
| 17 | Fzd4 | 1279 | 0.436 | 0.1198 | No |
| 18 | Coq5 | 1588 | 0.396 | 0.1092 | No |
| 19 | Retsat | 1630 | 0.392 | 0.1147 | No |
| 20 | Samm50 | 1736 | 0.379 | 0.1161 | No |
| 21 | Acly | 1795 | 0.374 | 0.1202 | No |
| 22 | Nabp1 | 1889 | 0.362 | 0.1219 | No |
| 23 | Ubqln1 | 1909 | 0.359 | 0.1280 | No |
| 24 | Ccng2 | 1984 | 0.352 | 0.1307 | No |
| 25 | Uqcr10 | 2009 | 0.350 | 0.1364 | No |
| 26 | Sult1a1 | 2028 | 0.348 | 0.1424 | No |
| 27 | Uqcrq | 2125 | 0.341 | 0.1435 | No |
| 28 | Atl2 | 2215 | 0.332 | 0.1449 | No |
| 29 | C3 | 2968 | 0.275 | 0.1049 | No |
| 30 | Acadm | 3089 | 0.268 | 0.1031 | No |
| 31 | Ywhag | 3100 | 0.267 | 0.1079 | No |
| 32 | Adcy6 | 3155 | 0.262 | 0.1100 | No |
| 33 | Idh3a | 3425 | 0.245 | 0.0987 | No |
| 34 | Scp2 | 3448 | 0.243 | 0.1023 | No |
| 35 | Dnajb9 | 3671 | 0.227 | 0.0935 | No |
| 36 | Cpt2 | 3683 | 0.227 | 0.0975 | No |
| 37 | Reep6 | 3823 | 0.217 | 0.0935 | No |
| 38 | Uqcrc1 | 3919 | 0.211 | 0.0920 | No |
| 39 | Slc27a1 | 4067 | 0.203 | 0.0872 | No |
| 40 | Ucp2 | 4099 | 0.201 | 0.0894 | No |
| 41 | Aldoa | 4142 | 0.198 | 0.0909 | No |
| 42 | Suclg1 | 4252 | 0.190 | 0.0882 | No |
| 43 | Rreb1 | 4426 | 0.180 | 0.0814 | No |
| 44 | Aldh2 | 4508 | 0.176 | 0.0801 | No |
| 45 | Pfkl | 4614 | 0.169 | 0.0771 | No |
| 46 | Ndufb7 | 4726 | 0.163 | 0.0738 | No |
| 47 | Mgll | 4865 | 0.156 | 0.0686 | No |
| 48 | Esyt1 | 4871 | 0.156 | 0.0714 | No |
| 49 | Apoe | 4890 | 0.154 | 0.0735 | No |
| 50 | Gpd2 | 4895 | 0.154 | 0.0764 | No |
| 51 | Ak2 | 4929 | 0.152 | 0.0775 | No |
| 52 | Grpel1 | 5598 | 0.115 | 0.0394 | No |
| 53 | Dhcr7 | 5708 | 0.113 | 0.0351 | No |
| 54 | Arl4a | 5717 | 0.113 | 0.0369 | No |
| 55 | G3bp2 | 5863 | 0.104 | 0.0302 | No |
| 56 | Pex14 | 5881 | 0.103 | 0.0313 | No |
| 57 | Scarb1 | 5891 | 0.102 | 0.0328 | No |
| 58 | Rnf11 | 5997 | 0.096 | 0.0285 | No |
| 59 | Idh1 | 6045 | 0.094 | 0.0275 | No |
| 60 | Gbe1 | 6083 | 0.092 | 0.0271 | No |
| 61 | Rmdn3 | 6094 | 0.092 | 0.0284 | No |
| 62 | Ptcd3 | 6298 | 0.080 | 0.0177 | No |
| 63 | Crat | 6429 | 0.073 | 0.0114 | No |
| 64 | Coq9 | 6459 | 0.071 | 0.0111 | No |
| 65 | Elovl6 | 6475 | 0.071 | 0.0116 | No |
| 66 | Lipe | 6600 | 0.064 | 0.0054 | No |
| 67 | Sdhb | 6610 | 0.064 | 0.0061 | No |
| 68 | Preb | 6664 | 0.061 | 0.0042 | No |
| 69 | Miga2 | 6736 | 0.057 | 0.0010 | No |
| 70 | Acadl | 6822 | 0.053 | -0.0030 | No |
| 71 | Mrpl15 | 7034 | 0.042 | -0.0149 | No |
| 72 | Hibch | 7114 | 0.038 | -0.0189 | No |
| 73 | Cavin1 | 7193 | 0.035 | -0.0230 | No |
| 74 | Chchd10 | 7266 | 0.032 | -0.0267 | No |
| 75 | Lpcat3 | 7363 | 0.028 | -0.0319 | No |
| 76 | Pdcd4 | 7485 | 0.022 | -0.0388 | No |
| 77 | Reep5 | 7557 | 0.018 | -0.0427 | No |
| 78 | Dhrs7 | 7713 | 0.011 | -0.0519 | No |
| 79 | Pemt | 7754 | 0.008 | -0.0541 | No |
| 80 | Bcl2l13 | 7810 | 0.006 | -0.0573 | No |
| 81 | Sdhc | 7846 | 0.004 | -0.0594 | No |
| 82 | Cd36 | 7928 | 0.000 | -0.0643 | No |
| 83 | Dram2 | 7999 | -0.001 | -0.0685 | No |
| 84 | Esrra | 8003 | -0.001 | -0.0687 | No |
| 85 | Ephx2 | 8119 | -0.007 | -0.0755 | No |
| 86 | Coq3 | 8216 | -0.012 | -0.0810 | No |
| 87 | Etfb | 8242 | -0.013 | -0.0823 | No |
| 88 | Dhrs7b | 8250 | -0.014 | -0.0824 | No |
| 89 | Jagn1 | 8388 | -0.020 | -0.0903 | No |
| 90 | Pfkfb3 | 8530 | -0.026 | -0.0983 | No |
| 91 | Adipor2 | 8641 | -0.032 | -0.1043 | No |
| 92 | Tank | 8653 | -0.033 | -0.1043 | No |
| 93 | Riok3 | 8790 | -0.040 | -0.1118 | No |
| 94 | Ptger3 | 9016 | -0.052 | -0.1243 | No |
| 95 | Angpt1 | 9082 | -0.055 | -0.1271 | No |
| 96 | Dld | 9273 | -0.065 | -0.1373 | No |
| 97 | Acox1 | 9365 | -0.071 | -0.1414 | No |
| 98 | Nmt1 | 9474 | -0.076 | -0.1463 | No |
| 99 | Decr1 | 9633 | -0.085 | -0.1542 | No |
| 100 | Uqcr11 | 9694 | -0.088 | -0.1560 | No |
| 101 | Gphn | 9771 | -0.092 | -0.1587 | No |
| 102 | Gpat4 | 10138 | -0.109 | -0.1787 | No |
| 103 | Dlat | 10205 | -0.113 | -0.1804 | No |
| 104 | Prdx3 | 10249 | -0.115 | -0.1806 | No |
| 105 | Omd | 10289 | -0.117 | -0.1806 | No |
| 106 | Ubc | 10310 | -0.118 | -0.1794 | No |
| 107 | Hadh | 10311 | -0.118 | -0.1770 | No |
| 108 | Cavin2 | 10324 | -0.119 | -0.1754 | No |
| 109 | Ifngr1 | 10742 | -0.141 | -0.1977 | No |
| 110 | Ppm1b | 10822 | -0.145 | -0.1995 | No |
| 111 | Itsn1 | 11229 | -0.166 | -0.2207 | No |
| 112 | Lpl | 11252 | -0.167 | -0.2187 | No |
| 113 | Aifm1 | 11376 | -0.175 | -0.2226 | No |
| 114 | Araf | 11385 | -0.175 | -0.2195 | No |
| 115 | Qdpr | 11531 | -0.183 | -0.2245 | No |
| 116 | Tob1 | 11669 | -0.190 | -0.2290 | No |
| 117 | Ndufa5 | 11710 | -0.192 | -0.2275 | No |
| 118 | Dnajc15 | 11859 | -0.201 | -0.2323 | No |
| 119 | Phyh | 12156 | -0.217 | -0.2458 | No |
| 120 | Sowahc | 12251 | -0.224 | -0.2470 | No |
| 121 | Ghitm | 12283 | -0.226 | -0.2443 | No |
| 122 | Por | 12294 | -0.227 | -0.2403 | No |
| 123 | Acads | 12469 | -0.238 | -0.2459 | No |
| 124 | Pim3 | 12718 | -0.251 | -0.2559 | No |
| 125 | Rtn3 | 12954 | -0.264 | -0.2647 | Yes |
| 126 | Atp1b3 | 12996 | -0.267 | -0.2618 | Yes |
| 127 | Stom | 12997 | -0.267 | -0.2563 | Yes |
| 128 | Plin2 | 13009 | -0.268 | -0.2515 | Yes |
| 129 | Tkt | 13049 | -0.271 | -0.2484 | Yes |
| 130 | Cox6a1 | 13104 | -0.274 | -0.2461 | Yes |
| 131 | Dgat1 | 13201 | -0.281 | -0.2462 | Yes |
| 132 | Vegfb | 13300 | -0.288 | -0.2463 | Yes |
| 133 | Slc1a5 | 13337 | -0.291 | -0.2425 | Yes |
| 134 | Agpat3 | 13442 | -0.298 | -0.2428 | Yes |
| 135 | Dbt | 13510 | -0.304 | -0.2406 | Yes |
| 136 | Baz2a | 13654 | -0.313 | -0.2429 | Yes |
| 137 | Echs1 | 13663 | -0.313 | -0.2370 | Yes |
| 138 | Idh3g | 13666 | -0.314 | -0.2308 | Yes |
| 139 | Lifr | 13723 | -0.317 | -0.2277 | Yes |
| 140 | Cd151 | 13877 | -0.328 | -0.2303 | Yes |
| 141 | Slc19a1 | 14214 | -0.353 | -0.2435 | Yes |
| 142 | Aco2 | 14216 | -0.353 | -0.2363 | Yes |
| 143 | Itih5 | 14416 | -0.373 | -0.2408 | Yes |
| 144 | Uck1 | 14425 | -0.374 | -0.2337 | Yes |
| 145 | Slc25a1 | 14536 | -0.386 | -0.2325 | Yes |
| 146 | Map4k3 | 14570 | -0.389 | -0.2266 | Yes |
| 147 | Gpx4 | 14577 | -0.390 | -0.2190 | Yes |
| 148 | Mgst3 | 14650 | -0.396 | -0.2153 | Yes |
| 149 | Gpx3 | 14771 | -0.406 | -0.2143 | Yes |
| 150 | Ddt | 14865 | -0.414 | -0.2115 | Yes |
| 151 | Col4a1 | 15000 | -0.430 | -0.2109 | Yes |
| 152 | Cat | 15032 | -0.434 | -0.2039 | Yes |
| 153 | Mtarc2 | 15116 | -0.445 | -0.1999 | Yes |
| 154 | Cyc1 | 15277 | -0.466 | -0.2001 | Yes |
| 155 | Acaa2 | 15303 | -0.470 | -0.1921 | Yes |
| 156 | Ltc4s | 15356 | -0.478 | -0.1855 | Yes |
| 157 | Bckdha | 15400 | -0.483 | -0.1783 | Yes |
| 158 | Slc25a10 | 15415 | -0.485 | -0.1692 | Yes |
| 159 | Cs | 15442 | -0.487 | -0.1609 | Yes |
| 160 | Abca1 | 15472 | -0.492 | -0.1527 | Yes |
| 161 | Ech1 | 15487 | -0.494 | -0.1435 | Yes |
| 162 | Sqor | 15656 | -0.523 | -0.1430 | Yes |
| 163 | Cox8a | 15668 | -0.525 | -0.1330 | Yes |
| 164 | Angptl4 | 15680 | -0.528 | -0.1229 | Yes |
| 165 | Abcb8 | 15716 | -0.534 | -0.1141 | Yes |
| 166 | Mtch2 | 15790 | -0.548 | -0.1074 | Yes |
| 167 | Cd302 | 15940 | -0.571 | -0.1048 | Yes |
| 168 | Mdh2 | 15950 | -0.573 | -0.0937 | Yes |
| 169 | Stat5a | 15976 | -0.578 | -0.0835 | Yes |
| 170 | Gadd45a | 15977 | -0.578 | -0.0717 | Yes |
| 171 | Taldo1 | 15984 | -0.579 | -0.0603 | Yes |
| 172 | Ndufab1 | 16040 | -0.593 | -0.0515 | Yes |
| 173 | Cmpk1 | 16156 | -0.625 | -0.0458 | Yes |
| 174 | Cox7b | 16240 | -0.648 | -0.0376 | Yes |
| 175 | Sod1 | 16651 | -0.977 | -0.0426 | Yes |
| 176 | Mylk | 16668 | -1.055 | -0.0221 | Yes |
| 177 | Ndufs3 | 16690 | -1.175 | 0.0005 | Yes |