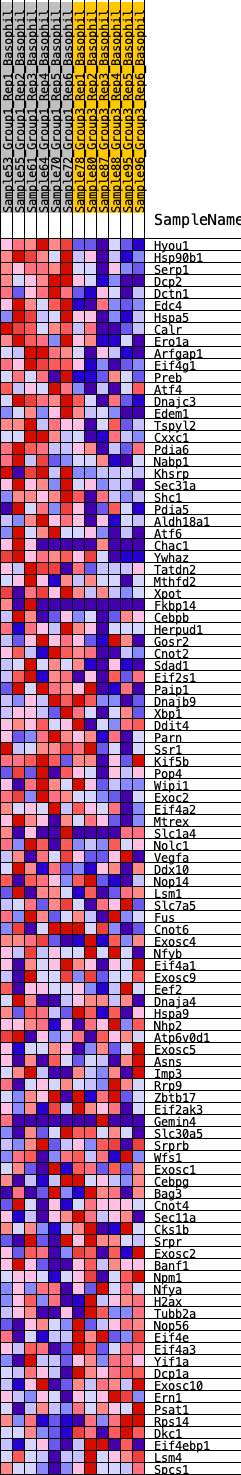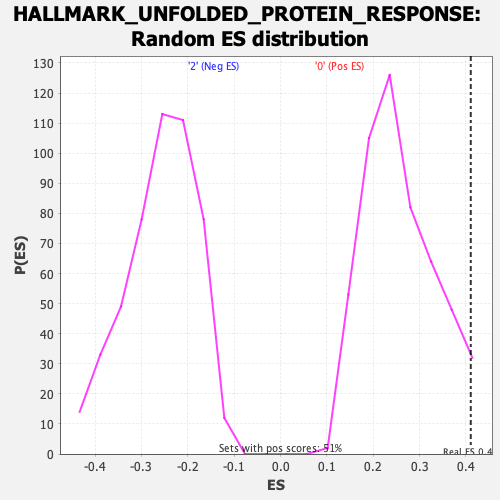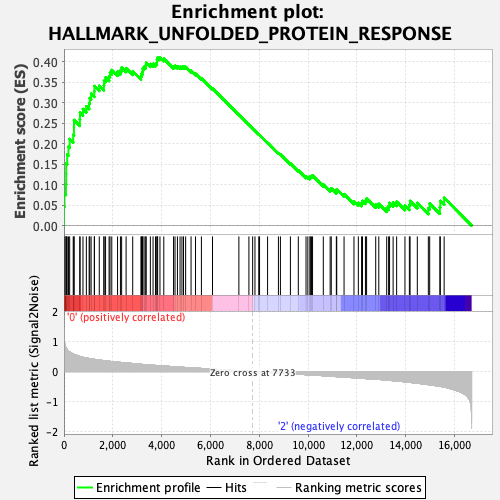
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.41032246 |
| Normalized Enrichment Score (NES) | 1.5789273 |
| Nominal p-value | 0.03515625 |
| FDR q-value | 0.1086876 |
| FWER p-Value | 0.226 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hyou1 | 2 | 1.428 | 0.0469 | Yes |
| 2 | Hsp90b1 | 20 | 0.972 | 0.0779 | Yes |
| 3 | Serp1 | 79 | 0.804 | 0.1009 | Yes |
| 4 | Dcp2 | 91 | 0.782 | 0.1260 | Yes |
| 5 | Dctn1 | 92 | 0.781 | 0.1517 | Yes |
| 6 | Edc4 | 130 | 0.729 | 0.1734 | Yes |
| 7 | Hspa5 | 183 | 0.677 | 0.1926 | Yes |
| 8 | Calr | 232 | 0.652 | 0.2112 | Yes |
| 9 | Ero1a | 378 | 0.584 | 0.2217 | Yes |
| 10 | Arfgap1 | 408 | 0.573 | 0.2388 | Yes |
| 11 | Eif4g1 | 410 | 0.572 | 0.2576 | Yes |
| 12 | Preb | 649 | 0.504 | 0.2598 | Yes |
| 13 | Atf4 | 656 | 0.503 | 0.2760 | Yes |
| 14 | Dnajc3 | 777 | 0.475 | 0.2844 | Yes |
| 15 | Edem1 | 916 | 0.454 | 0.2910 | Yes |
| 16 | Tspyl2 | 1030 | 0.435 | 0.2985 | Yes |
| 17 | Cxxc1 | 1056 | 0.429 | 0.3112 | Yes |
| 18 | Pdia6 | 1111 | 0.422 | 0.3218 | Yes |
| 19 | Nabp1 | 1242 | 0.407 | 0.3274 | Yes |
| 20 | Khsrp | 1247 | 0.406 | 0.3405 | Yes |
| 21 | Sec31a | 1445 | 0.383 | 0.3412 | Yes |
| 22 | Shc1 | 1633 | 0.363 | 0.3419 | Yes |
| 23 | Pdia5 | 1641 | 0.362 | 0.3534 | Yes |
| 24 | Aldh18a1 | 1703 | 0.355 | 0.3614 | Yes |
| 25 | Atf6 | 1847 | 0.342 | 0.3640 | Yes |
| 26 | Chac1 | 1885 | 0.338 | 0.3729 | Yes |
| 27 | Ywhaz | 1957 | 0.331 | 0.3796 | Yes |
| 28 | Tatdn2 | 2194 | 0.312 | 0.3757 | Yes |
| 29 | Mthfd2 | 2312 | 0.303 | 0.3786 | Yes |
| 30 | Xpot | 2361 | 0.299 | 0.3855 | Yes |
| 31 | Fkbp14 | 2544 | 0.285 | 0.3840 | Yes |
| 32 | Cebpb | 2815 | 0.267 | 0.3765 | Yes |
| 33 | Herpud1 | 3152 | 0.240 | 0.3641 | Yes |
| 34 | Gosr2 | 3177 | 0.239 | 0.3705 | Yes |
| 35 | Cnot2 | 3217 | 0.235 | 0.3759 | Yes |
| 36 | Sdad1 | 3226 | 0.235 | 0.3832 | Yes |
| 37 | Eif2s1 | 3281 | 0.232 | 0.3876 | Yes |
| 38 | Paip1 | 3348 | 0.227 | 0.3911 | Yes |
| 39 | Dnajb9 | 3366 | 0.225 | 0.3974 | Yes |
| 40 | Xbp1 | 3538 | 0.215 | 0.3942 | Yes |
| 41 | Ddit4 | 3648 | 0.208 | 0.3945 | Yes |
| 42 | Parn | 3748 | 0.202 | 0.3952 | Yes |
| 43 | Ssr1 | 3806 | 0.199 | 0.3983 | Yes |
| 44 | Kif5b | 3810 | 0.199 | 0.4047 | Yes |
| 45 | Pop4 | 3836 | 0.197 | 0.4097 | Yes |
| 46 | Wipi1 | 3931 | 0.192 | 0.4103 | Yes |
| 47 | Exoc2 | 4088 | 0.182 | 0.4069 | No |
| 48 | Eif4a2 | 4488 | 0.160 | 0.3882 | No |
| 49 | Mtrex | 4548 | 0.157 | 0.3898 | No |
| 50 | Slc1a4 | 4647 | 0.151 | 0.3888 | No |
| 51 | Nolc1 | 4754 | 0.145 | 0.3872 | No |
| 52 | Vegfa | 4832 | 0.141 | 0.3873 | No |
| 53 | Ddx10 | 4896 | 0.138 | 0.3880 | No |
| 54 | Nop14 | 4983 | 0.133 | 0.3872 | No |
| 55 | Lsm1 | 5205 | 0.121 | 0.3779 | No |
| 56 | Slc7a5 | 5388 | 0.114 | 0.3706 | No |
| 57 | Fus | 5629 | 0.101 | 0.3595 | No |
| 58 | Cnot6 | 6084 | 0.079 | 0.3348 | No |
| 59 | Exosc4 | 7164 | 0.025 | 0.2706 | No |
| 60 | Nfyb | 7575 | 0.006 | 0.2461 | No |
| 61 | Eif4a1 | 7728 | 0.000 | 0.2369 | No |
| 62 | Exosc9 | 7823 | -0.003 | 0.2314 | No |
| 63 | Eef2 | 7979 | -0.010 | 0.2223 | No |
| 64 | Dnaja4 | 8011 | -0.011 | 0.2208 | No |
| 65 | Hspa9 | 8340 | -0.027 | 0.2020 | No |
| 66 | Nhp2 | 8784 | -0.048 | 0.1769 | No |
| 67 | Atp6v0d1 | 8866 | -0.051 | 0.1737 | No |
| 68 | Exosc5 | 9275 | -0.072 | 0.1515 | No |
| 69 | Asns | 9599 | -0.087 | 0.1349 | No |
| 70 | Imp3 | 9915 | -0.104 | 0.1193 | No |
| 71 | Rrp9 | 9990 | -0.107 | 0.1184 | No |
| 72 | Zbtb17 | 10076 | -0.112 | 0.1169 | No |
| 73 | Eif2ak3 | 10090 | -0.113 | 0.1199 | No |
| 74 | Gemin4 | 10138 | -0.115 | 0.1208 | No |
| 75 | Slc30a5 | 10182 | -0.118 | 0.1221 | No |
| 76 | Srprb | 10623 | -0.138 | 0.1001 | No |
| 77 | Wfs1 | 10904 | -0.154 | 0.0883 | No |
| 78 | Exosc1 | 10949 | -0.157 | 0.0909 | No |
| 79 | Cebpg | 11157 | -0.168 | 0.0839 | No |
| 80 | Bag3 | 11173 | -0.169 | 0.0886 | No |
| 81 | Cnot4 | 11473 | -0.184 | 0.0766 | No |
| 82 | Sec11a | 11879 | -0.207 | 0.0590 | No |
| 83 | Cks1b | 12058 | -0.217 | 0.0555 | No |
| 84 | Srpr | 12191 | -0.225 | 0.0549 | No |
| 85 | Exosc2 | 12229 | -0.228 | 0.0602 | No |
| 86 | Banf1 | 12349 | -0.234 | 0.0607 | No |
| 87 | Npm1 | 12396 | -0.236 | 0.0657 | No |
| 88 | Nfya | 12770 | -0.257 | 0.0517 | No |
| 89 | H2ax | 12896 | -0.264 | 0.0529 | No |
| 90 | Tubb2a | 13220 | -0.284 | 0.0428 | No |
| 91 | Nop56 | 13299 | -0.291 | 0.0476 | No |
| 92 | Eif4e | 13337 | -0.293 | 0.0551 | No |
| 93 | Eif4a3 | 13483 | -0.305 | 0.0564 | No |
| 94 | Yif1a | 13627 | -0.315 | 0.0581 | No |
| 95 | Dcp1a | 13968 | -0.343 | 0.0489 | No |
| 96 | Exosc10 | 14149 | -0.359 | 0.0499 | No |
| 97 | Ern1 | 14178 | -0.361 | 0.0601 | No |
| 98 | Psat1 | 14474 | -0.389 | 0.0551 | No |
| 99 | Rps14 | 14926 | -0.437 | 0.0424 | No |
| 100 | Dkc1 | 14981 | -0.443 | 0.0537 | No |
| 101 | Eif4ebp1 | 15398 | -0.493 | 0.0449 | No |
| 102 | Lsm4 | 15416 | -0.494 | 0.0601 | No |
| 103 | Spcs1 | 15573 | -0.520 | 0.0678 | No |