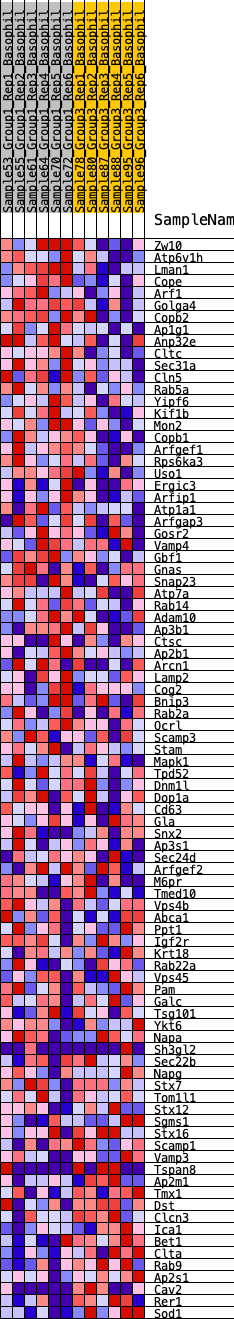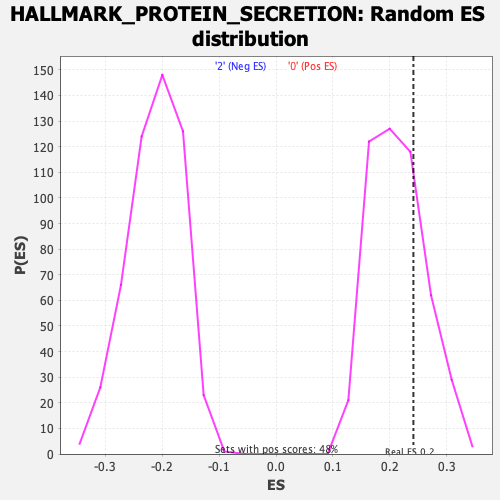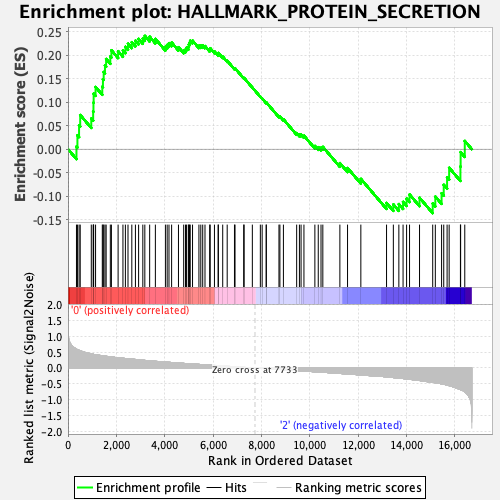
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.24110827 |
| Normalized Enrichment Score (NES) | 1.1365775 |
| Nominal p-value | 0.2634855 |
| FDR q-value | 0.5756545 |
| FWER p-Value | 0.957 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zw10 | 350 | 0.592 | 0.0057 | Yes |
| 2 | Atp6v1h | 390 | 0.580 | 0.0296 | Yes |
| 3 | Lman1 | 458 | 0.557 | 0.0508 | Yes |
| 4 | Cope | 505 | 0.545 | 0.0727 | Yes |
| 5 | Arf1 | 963 | 0.446 | 0.0654 | Yes |
| 6 | Golga4 | 1041 | 0.433 | 0.0804 | Yes |
| 7 | Copb2 | 1053 | 0.430 | 0.0992 | Yes |
| 8 | Ap1g1 | 1061 | 0.429 | 0.1181 | Yes |
| 9 | Anp32e | 1140 | 0.418 | 0.1324 | Yes |
| 10 | Cltc | 1418 | 0.386 | 0.1331 | Yes |
| 11 | Sec31a | 1445 | 0.383 | 0.1489 | Yes |
| 12 | Cln5 | 1476 | 0.379 | 0.1643 | Yes |
| 13 | Rab5a | 1529 | 0.375 | 0.1781 | Yes |
| 14 | Yipf6 | 1577 | 0.368 | 0.1920 | Yes |
| 15 | Kif1b | 1750 | 0.351 | 0.1975 | Yes |
| 16 | Mon2 | 1795 | 0.345 | 0.2105 | Yes |
| 17 | Copb1 | 2072 | 0.323 | 0.2085 | Yes |
| 18 | Arfgef1 | 2272 | 0.306 | 0.2104 | Yes |
| 19 | Rps6ka3 | 2373 | 0.298 | 0.2178 | Yes |
| 20 | Uso1 | 2481 | 0.289 | 0.2245 | Yes |
| 21 | Ergic3 | 2639 | 0.279 | 0.2277 | Yes |
| 22 | Arfip1 | 2788 | 0.269 | 0.2309 | Yes |
| 23 | Atp1a1 | 2920 | 0.256 | 0.2346 | Yes |
| 24 | Arfgap3 | 3094 | 0.244 | 0.2352 | Yes |
| 25 | Gosr2 | 3177 | 0.239 | 0.2411 | Yes |
| 26 | Vamp4 | 3377 | 0.225 | 0.2393 | No |
| 27 | Gbf1 | 3613 | 0.210 | 0.2346 | No |
| 28 | Gnas | 4028 | 0.186 | 0.2182 | No |
| 29 | Snap23 | 4101 | 0.181 | 0.2220 | No |
| 30 | Atp7a | 4176 | 0.177 | 0.2256 | No |
| 31 | Rab14 | 4285 | 0.172 | 0.2269 | No |
| 32 | Adam10 | 4568 | 0.156 | 0.2169 | No |
| 33 | Ap3b1 | 4784 | 0.144 | 0.2105 | No |
| 34 | Ctsc | 4860 | 0.140 | 0.2123 | No |
| 35 | Ap2b1 | 4903 | 0.138 | 0.2160 | No |
| 36 | Arcn1 | 4977 | 0.133 | 0.2177 | No |
| 37 | Lamp2 | 4995 | 0.133 | 0.2227 | No |
| 38 | Cog2 | 5026 | 0.131 | 0.2268 | No |
| 39 | Bnip3 | 5054 | 0.129 | 0.2310 | No |
| 40 | Rab2a | 5150 | 0.124 | 0.2309 | No |
| 41 | Ocrl | 5414 | 0.112 | 0.2202 | No |
| 42 | Scamp3 | 5492 | 0.109 | 0.2205 | No |
| 43 | Stam | 5571 | 0.105 | 0.2205 | No |
| 44 | Mapk1 | 5665 | 0.099 | 0.2194 | No |
| 45 | Tpd52 | 5859 | 0.091 | 0.2119 | No |
| 46 | Dnm1l | 5880 | 0.089 | 0.2147 | No |
| 47 | Dop1a | 6056 | 0.081 | 0.2078 | No |
| 48 | Cd63 | 6209 | 0.073 | 0.2020 | No |
| 49 | Gla | 6218 | 0.072 | 0.2048 | No |
| 50 | Snx2 | 6398 | 0.063 | 0.1968 | No |
| 51 | Ap3s1 | 6583 | 0.053 | 0.1881 | No |
| 52 | Sec24d | 6884 | 0.039 | 0.1718 | No |
| 53 | Arfgef2 | 6907 | 0.037 | 0.1722 | No |
| 54 | M6pr | 7266 | 0.020 | 0.1515 | No |
| 55 | Tmed10 | 7283 | 0.019 | 0.1514 | No |
| 56 | Vps4b | 7623 | 0.004 | 0.1312 | No |
| 57 | Abca1 | 7954 | -0.008 | 0.1117 | No |
| 58 | Ppt1 | 8029 | -0.012 | 0.1078 | No |
| 59 | Igf2r | 8198 | -0.020 | 0.0986 | No |
| 60 | Krt18 | 8199 | -0.020 | 0.0996 | No |
| 61 | Rab22a | 8724 | -0.045 | 0.0700 | No |
| 62 | Vps45 | 8763 | -0.046 | 0.0698 | No |
| 63 | Pam | 8908 | -0.054 | 0.0636 | No |
| 64 | Galc | 9456 | -0.081 | 0.0343 | No |
| 65 | Tsg101 | 9570 | -0.086 | 0.0314 | No |
| 66 | Ykt6 | 9638 | -0.089 | 0.0314 | No |
| 67 | Napa | 9757 | -0.096 | 0.0287 | No |
| 68 | Sh3gl2 | 10209 | -0.118 | 0.0069 | No |
| 69 | Sec22b | 10349 | -0.125 | 0.0042 | No |
| 70 | Napg | 10463 | -0.131 | 0.0033 | No |
| 71 | Stx7 | 10538 | -0.134 | 0.0049 | No |
| 72 | Tom1l1 | 11243 | -0.173 | -0.0296 | No |
| 73 | Stx12 | 11560 | -0.190 | -0.0401 | No |
| 74 | Sgms1 | 12110 | -0.220 | -0.0632 | No |
| 75 | Stx16 | 13173 | -0.281 | -0.1144 | No |
| 76 | Scamp1 | 13454 | -0.302 | -0.1176 | No |
| 77 | Vamp3 | 13682 | -0.320 | -0.1168 | No |
| 78 | Tspan8 | 13856 | -0.333 | -0.1121 | No |
| 79 | Ap2m1 | 14002 | -0.345 | -0.1052 | No |
| 80 | Tmx1 | 14124 | -0.356 | -0.0964 | No |
| 81 | Dst | 14537 | -0.395 | -0.1033 | No |
| 82 | Clcn3 | 15082 | -0.454 | -0.1155 | No |
| 83 | Ica1 | 15190 | -0.467 | -0.1008 | No |
| 84 | Bet1 | 15449 | -0.499 | -0.0937 | No |
| 85 | Clta | 15538 | -0.512 | -0.0759 | No |
| 86 | Rab9 | 15677 | -0.539 | -0.0597 | No |
| 87 | Ap2s1 | 15762 | -0.558 | -0.0395 | No |
| 88 | Cav2 | 16228 | -0.680 | -0.0368 | No |
| 89 | Rer1 | 16237 | -0.684 | -0.0063 | No |
| 90 | Sod1 | 16408 | -0.752 | 0.0175 | No |