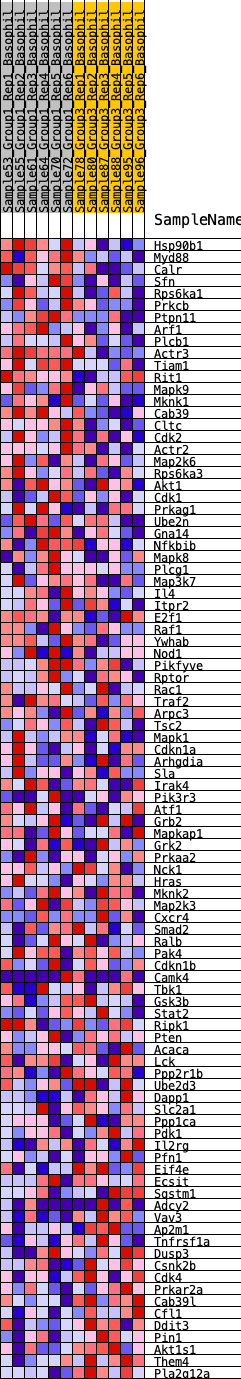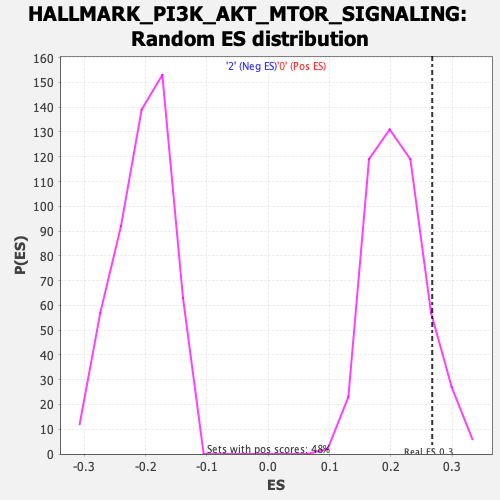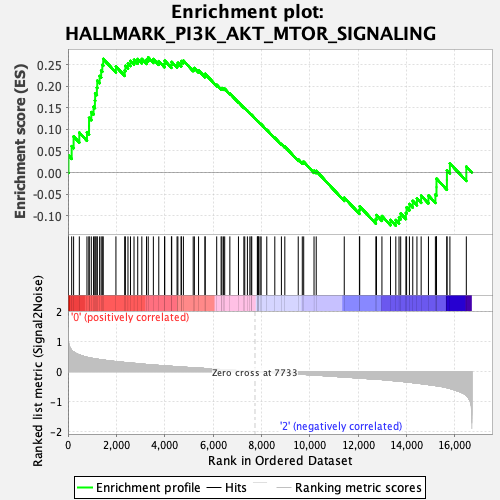
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.26743793 |
| Normalized Enrichment Score (NES) | 1.2736259 |
| Nominal p-value | 0.12603305 |
| FDR q-value | 0.3834355 |
| FWER p-Value | 0.842 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsp90b1 | 20 | 0.972 | 0.0395 | Yes |
| 2 | Myd88 | 152 | 0.705 | 0.0612 | Yes |
| 3 | Calr | 232 | 0.652 | 0.0837 | Yes |
| 4 | Sfn | 468 | 0.553 | 0.0927 | Yes |
| 5 | Rps6ka1 | 784 | 0.474 | 0.0936 | Yes |
| 6 | Prkcb | 870 | 0.461 | 0.1078 | Yes |
| 7 | Ptpn11 | 871 | 0.461 | 0.1271 | Yes |
| 8 | Arf1 | 963 | 0.446 | 0.1403 | Yes |
| 9 | Plcb1 | 1057 | 0.429 | 0.1527 | Yes |
| 10 | Actr3 | 1113 | 0.421 | 0.1670 | Yes |
| 11 | Tiam1 | 1123 | 0.420 | 0.1841 | Yes |
| 12 | Rit1 | 1194 | 0.412 | 0.1971 | Yes |
| 13 | Mapk9 | 1210 | 0.410 | 0.2134 | Yes |
| 14 | Mknk1 | 1308 | 0.398 | 0.2242 | Yes |
| 15 | Cab39 | 1371 | 0.390 | 0.2369 | Yes |
| 16 | Cltc | 1418 | 0.386 | 0.2503 | Yes |
| 17 | Cdk2 | 1457 | 0.381 | 0.2639 | Yes |
| 18 | Actr2 | 1982 | 0.330 | 0.2462 | Yes |
| 19 | Map2k6 | 2344 | 0.300 | 0.2371 | Yes |
| 20 | Rps6ka3 | 2373 | 0.298 | 0.2479 | Yes |
| 21 | Akt1 | 2480 | 0.289 | 0.2536 | Yes |
| 22 | Cdk1 | 2584 | 0.283 | 0.2593 | Yes |
| 23 | Prkag1 | 2730 | 0.272 | 0.2619 | Yes |
| 24 | Ube2n | 2883 | 0.260 | 0.2637 | Yes |
| 25 | Gna14 | 3053 | 0.246 | 0.2638 | Yes |
| 26 | Nfkbib | 3246 | 0.233 | 0.2621 | Yes |
| 27 | Mapk8 | 3317 | 0.229 | 0.2674 | Yes |
| 28 | Plcg1 | 3530 | 0.215 | 0.2637 | No |
| 29 | Map3k7 | 3754 | 0.202 | 0.2587 | No |
| 30 | Il4 | 3992 | 0.188 | 0.2523 | No |
| 31 | Itpr2 | 4001 | 0.188 | 0.2597 | No |
| 32 | E2f1 | 4277 | 0.172 | 0.2504 | No |
| 33 | Raf1 | 4289 | 0.171 | 0.2569 | No |
| 34 | Ywhab | 4510 | 0.159 | 0.2503 | No |
| 35 | Nod1 | 4544 | 0.158 | 0.2549 | No |
| 36 | Pikfyve | 4681 | 0.149 | 0.2530 | No |
| 37 | Rptor | 4685 | 0.149 | 0.2590 | No |
| 38 | Rac1 | 4772 | 0.145 | 0.2599 | No |
| 39 | Traf2 | 5177 | 0.123 | 0.2407 | No |
| 40 | Arpc3 | 5226 | 0.120 | 0.2428 | No |
| 41 | Tsc2 | 5399 | 0.113 | 0.2372 | No |
| 42 | Mapk1 | 5665 | 0.099 | 0.2254 | No |
| 43 | Cdkn1a | 5671 | 0.099 | 0.2293 | No |
| 44 | Arhgdia | 6149 | 0.076 | 0.2037 | No |
| 45 | Sla | 6332 | 0.066 | 0.1955 | No |
| 46 | Irak4 | 6367 | 0.064 | 0.1962 | No |
| 47 | Pik3r3 | 6434 | 0.061 | 0.1947 | No |
| 48 | Atf1 | 6480 | 0.058 | 0.1945 | No |
| 49 | Grb2 | 6693 | 0.048 | 0.1837 | No |
| 50 | Mapkap1 | 7049 | 0.030 | 0.1636 | No |
| 51 | Grk2 | 7273 | 0.019 | 0.1510 | No |
| 52 | Prkaa2 | 7310 | 0.018 | 0.1496 | No |
| 53 | Nck1 | 7419 | 0.012 | 0.1436 | No |
| 54 | Hras | 7520 | 0.008 | 0.1379 | No |
| 55 | Mknk2 | 7572 | 0.006 | 0.1351 | No |
| 56 | Map2k3 | 7599 | 0.005 | 0.1337 | No |
| 57 | Cxcr4 | 7830 | -0.003 | 0.1200 | No |
| 58 | Smad2 | 7848 | -0.003 | 0.1191 | No |
| 59 | Ralb | 7864 | -0.004 | 0.1184 | No |
| 60 | Pak4 | 7884 | -0.005 | 0.1175 | No |
| 61 | Cdkn1b | 7942 | -0.008 | 0.1144 | No |
| 62 | Camk4 | 7987 | -0.010 | 0.1121 | No |
| 63 | Tbk1 | 8220 | -0.021 | 0.0990 | No |
| 64 | Gsk3b | 8552 | -0.037 | 0.0807 | No |
| 65 | Stat2 | 8825 | -0.050 | 0.0664 | No |
| 66 | Ripk1 | 8966 | -0.056 | 0.0603 | No |
| 67 | Pten | 9521 | -0.084 | 0.0304 | No |
| 68 | Acaca | 9689 | -0.092 | 0.0242 | No |
| 69 | Lck | 9747 | -0.095 | 0.0248 | No |
| 70 | Ppp2r1b | 10174 | -0.117 | 0.0040 | No |
| 71 | Ube2d3 | 10265 | -0.121 | 0.0037 | No |
| 72 | Dapp1 | 11424 | -0.182 | -0.0585 | No |
| 73 | Slc2a1 | 12051 | -0.217 | -0.0871 | No |
| 74 | Ppp1ca | 12063 | -0.218 | -0.0786 | No |
| 75 | Pdk1 | 12731 | -0.254 | -0.1082 | No |
| 76 | Il2rg | 12757 | -0.256 | -0.0989 | No |
| 77 | Pfn1 | 12982 | -0.267 | -0.1012 | No |
| 78 | Eif4e | 13337 | -0.293 | -0.1103 | No |
| 79 | Ecsit | 13556 | -0.309 | -0.1105 | No |
| 80 | Sqstm1 | 13689 | -0.321 | -0.1050 | No |
| 81 | Adcy2 | 13758 | -0.327 | -0.0954 | No |
| 82 | Vav3 | 13974 | -0.343 | -0.0939 | No |
| 83 | Ap2m1 | 14002 | -0.345 | -0.0811 | No |
| 84 | Tnfrsf1a | 14119 | -0.355 | -0.0732 | No |
| 85 | Dusp3 | 14257 | -0.370 | -0.0660 | No |
| 86 | Csnk2b | 14430 | -0.386 | -0.0602 | No |
| 87 | Cdk4 | 14604 | -0.404 | -0.0537 | No |
| 88 | Prkar2a | 14906 | -0.436 | -0.0536 | No |
| 89 | Cab39l | 15193 | -0.467 | -0.0512 | No |
| 90 | Cfl1 | 15238 | -0.472 | -0.0341 | No |
| 91 | Ddit3 | 15239 | -0.472 | -0.0143 | No |
| 92 | Pin1 | 15669 | -0.538 | -0.0176 | No |
| 93 | Akt1s1 | 15670 | -0.538 | 0.0049 | No |
| 94 | Them4 | 15793 | -0.564 | 0.0212 | No |
| 95 | Pla2g12a | 16472 | -0.795 | 0.0137 | No |