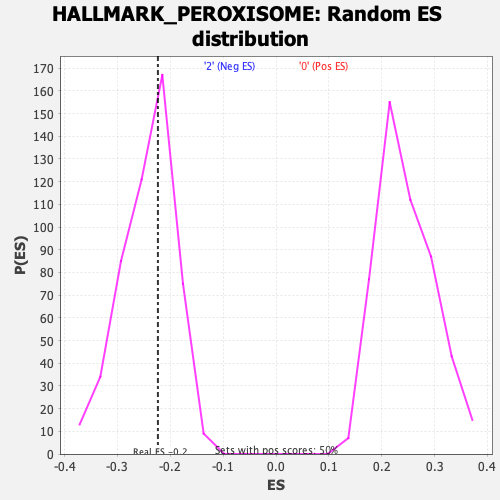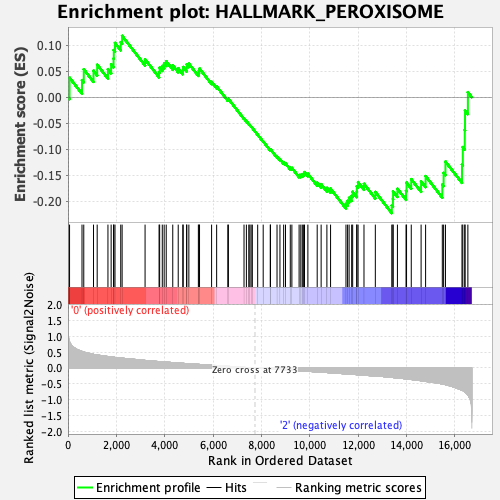
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.22338727 |
| Normalized Enrichment Score (NES) | -0.92477083 |
| Nominal p-value | 0.58928573 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcd2 | 64 | 0.830 | 0.0377 | No |
| 2 | Pex14 | 579 | 0.523 | 0.0330 | No |
| 3 | Itgb1bp1 | 652 | 0.504 | 0.0539 | No |
| 4 | Dhcr24 | 1055 | 0.430 | 0.0513 | No |
| 5 | Sema3c | 1207 | 0.411 | 0.0628 | No |
| 6 | Idh1 | 1652 | 0.361 | 0.0542 | No |
| 7 | Ide | 1784 | 0.346 | 0.0636 | No |
| 8 | Acsl4 | 1884 | 0.338 | 0.0746 | No |
| 9 | Msh2 | 1893 | 0.337 | 0.0911 | No |
| 10 | Ehhadh | 1943 | 0.332 | 0.1048 | No |
| 11 | Ywhah | 2181 | 0.313 | 0.1062 | No |
| 12 | Fads1 | 2238 | 0.309 | 0.1184 | No |
| 13 | Bcl10 | 3187 | 0.238 | 0.0732 | No |
| 14 | Cat | 3766 | 0.201 | 0.0485 | No |
| 15 | Scp2 | 3790 | 0.200 | 0.0572 | No |
| 16 | Top2a | 3899 | 0.194 | 0.0604 | No |
| 17 | Dhrs3 | 3977 | 0.189 | 0.0652 | No |
| 18 | Gstk1 | 4062 | 0.184 | 0.0694 | No |
| 19 | Isoc1 | 4332 | 0.169 | 0.0617 | No |
| 20 | Retsat | 4558 | 0.156 | 0.0560 | No |
| 21 | Slc23a2 | 4749 | 0.146 | 0.0518 | No |
| 22 | Crat | 4762 | 0.145 | 0.0584 | No |
| 23 | Abcd3 | 4908 | 0.137 | 0.0565 | No |
| 24 | Fdps | 4909 | 0.137 | 0.0634 | No |
| 25 | Acox1 | 4998 | 0.133 | 0.0648 | No |
| 26 | Pex5 | 5391 | 0.114 | 0.0469 | No |
| 27 | Eci2 | 5416 | 0.112 | 0.0511 | No |
| 28 | Aldh9a1 | 5439 | 0.111 | 0.0553 | No |
| 29 | Tspo | 5938 | 0.087 | 0.0297 | No |
| 30 | Ephx2 | 6147 | 0.076 | 0.0210 | No |
| 31 | Dlg4 | 6609 | 0.052 | -0.0042 | No |
| 32 | Smarcc1 | 6633 | 0.051 | -0.0030 | No |
| 33 | Pex6 | 7279 | 0.019 | -0.0409 | No |
| 34 | Slc35b2 | 7377 | 0.014 | -0.0460 | No |
| 35 | Acsl5 | 7476 | 0.010 | -0.0514 | No |
| 36 | Hras | 7520 | 0.008 | -0.0536 | No |
| 37 | Cnbp | 7573 | 0.006 | -0.0564 | No |
| 38 | Vps4b | 7623 | 0.004 | -0.0591 | No |
| 39 | Acsl1 | 7844 | -0.003 | -0.0722 | No |
| 40 | Pex2 | 8072 | -0.015 | -0.0851 | No |
| 41 | Abcc5 | 8364 | -0.028 | -0.1012 | No |
| 42 | Slc25a17 | 8374 | -0.029 | -0.1003 | No |
| 43 | Hsd17b4 | 8644 | -0.042 | -0.1144 | No |
| 44 | Acot8 | 8765 | -0.047 | -0.1193 | No |
| 45 | Pabpc1 | 8918 | -0.054 | -0.1257 | No |
| 46 | Idh2 | 8994 | -0.058 | -0.1273 | No |
| 47 | Lonp2 | 9192 | -0.067 | -0.1358 | No |
| 48 | Prdx5 | 9258 | -0.071 | -0.1362 | No |
| 49 | Ctbp1 | 9557 | -0.086 | -0.1498 | No |
| 50 | Cdk7 | 9628 | -0.089 | -0.1496 | No |
| 51 | Hsd17b11 | 9690 | -0.092 | -0.1486 | No |
| 52 | Mlycd | 9745 | -0.095 | -0.1471 | No |
| 53 | Cln8 | 9780 | -0.097 | -0.1443 | No |
| 54 | Ercc3 | 9920 | -0.104 | -0.1475 | No |
| 55 | Nudt19 | 10305 | -0.123 | -0.1644 | No |
| 56 | Cacna1b | 10468 | -0.131 | -0.1676 | No |
| 57 | Sod2 | 10706 | -0.143 | -0.1747 | No |
| 58 | Slc25a19 | 10858 | -0.152 | -0.1762 | No |
| 59 | Idi1 | 11495 | -0.186 | -0.2051 | No |
| 60 | Gnpat | 11562 | -0.190 | -0.1996 | No |
| 61 | Acaa1a | 11625 | -0.193 | -0.1936 | No |
| 62 | Hmgcl | 11731 | -0.199 | -0.1900 | No |
| 63 | Hsd3b7 | 11772 | -0.201 | -0.1823 | No |
| 64 | Sult2b1 | 11941 | -0.211 | -0.1819 | No |
| 65 | Abcd1 | 11945 | -0.211 | -0.1715 | No |
| 66 | Elovl5 | 11996 | -0.214 | -0.1638 | No |
| 67 | Rdh11 | 12240 | -0.228 | -0.1669 | No |
| 68 | Pex11b | 12709 | -0.252 | -0.1825 | No |
| 69 | Ercc1 | 13390 | -0.297 | -0.2085 | Yes |
| 70 | Mvp | 13437 | -0.301 | -0.1962 | Yes |
| 71 | Atxn1 | 13444 | -0.301 | -0.1814 | Yes |
| 72 | Abcb9 | 13623 | -0.315 | -0.1764 | Yes |
| 73 | Prdx1 | 13979 | -0.344 | -0.1805 | Yes |
| 74 | Aldh1a1 | 14000 | -0.345 | -0.1644 | Yes |
| 75 | Cln6 | 14195 | -0.363 | -0.1579 | Yes |
| 76 | Pex11a | 14602 | -0.404 | -0.1621 | Yes |
| 77 | Slc25a4 | 14788 | -0.424 | -0.1520 | Yes |
| 78 | Cadm1 | 15479 | -0.504 | -0.1682 | Yes |
| 79 | Abcb1a | 15533 | -0.511 | -0.1458 | Yes |
| 80 | Pex13 | 15609 | -0.526 | -0.1239 | Yes |
| 81 | Siah1a | 16293 | -0.705 | -0.1297 | Yes |
| 82 | Ech1 | 16327 | -0.713 | -0.0960 | Yes |
| 83 | Sod1 | 16408 | -0.752 | -0.0631 | Yes |
| 84 | Abcb4 | 16419 | -0.759 | -0.0256 | Yes |
| 85 | Fis1 | 16536 | -0.846 | 0.0098 | Yes |