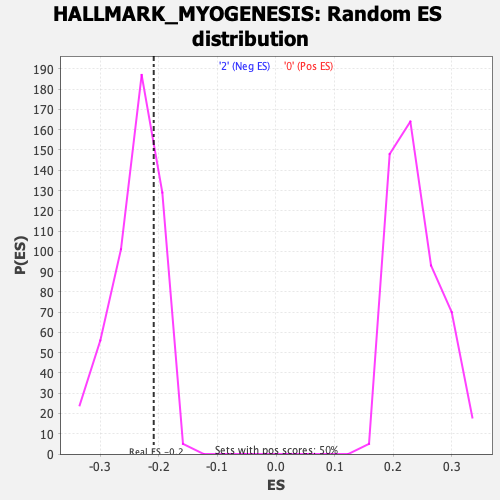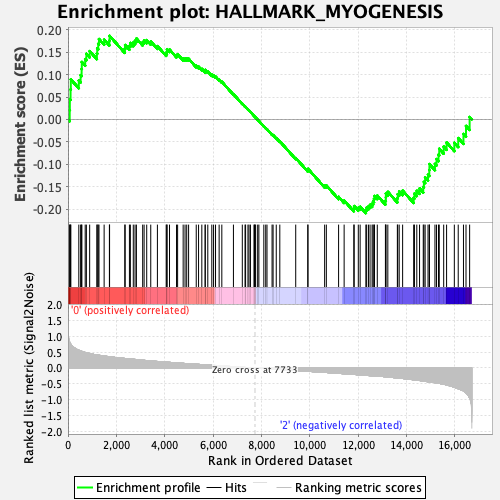
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.20889522 |
| Normalized Enrichment Score (NES) | -0.8709274 |
| Nominal p-value | 0.75697213 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnnt1 | 70 | 0.814 | 0.0207 | No |
| 2 | Lpin1 | 76 | 0.808 | 0.0452 | No |
| 3 | Ifrd1 | 106 | 0.764 | 0.0669 | No |
| 4 | Gsn | 116 | 0.751 | 0.0894 | No |
| 5 | Flii | 447 | 0.561 | 0.0867 | No |
| 6 | Dapk2 | 522 | 0.539 | 0.0987 | No |
| 7 | Myl6b | 563 | 0.526 | 0.1124 | No |
| 8 | Dmpk | 571 | 0.525 | 0.1281 | No |
| 9 | Notch1 | 711 | 0.488 | 0.1347 | No |
| 10 | Mylpf | 760 | 0.478 | 0.1464 | No |
| 11 | Myh9 | 896 | 0.457 | 0.1523 | No |
| 12 | Rit1 | 1194 | 0.412 | 0.1470 | No |
| 13 | Itgb4 | 1209 | 0.411 | 0.1587 | No |
| 14 | Rb1 | 1250 | 0.405 | 0.1688 | No |
| 15 | Hdac5 | 1281 | 0.401 | 0.1793 | No |
| 16 | Klf5 | 1494 | 0.378 | 0.1781 | No |
| 17 | Kcnh1 | 1712 | 0.355 | 0.1758 | No |
| 18 | Pick1 | 1719 | 0.354 | 0.1863 | No |
| 19 | Myo1c | 2349 | 0.300 | 0.1576 | No |
| 20 | Itgb1 | 2363 | 0.299 | 0.1660 | No |
| 21 | Nos1 | 2540 | 0.285 | 0.1641 | No |
| 22 | Sptan1 | 2579 | 0.284 | 0.1705 | No |
| 23 | Pde4dip | 2700 | 0.274 | 0.1717 | No |
| 24 | Agl | 2772 | 0.270 | 0.1757 | No |
| 25 | Gabarapl2 | 2829 | 0.266 | 0.1804 | No |
| 26 | Itgb5 | 3088 | 0.244 | 0.1724 | No |
| 27 | Reep1 | 3147 | 0.240 | 0.1762 | No |
| 28 | Hbegf | 3255 | 0.233 | 0.1769 | No |
| 29 | Camk2b | 3422 | 0.222 | 0.1737 | No |
| 30 | Col4a2 | 3697 | 0.205 | 0.1635 | No |
| 31 | Mef2a | 4060 | 0.184 | 0.1473 | No |
| 32 | Agrn | 4089 | 0.182 | 0.1512 | No |
| 33 | Ocel1 | 4094 | 0.182 | 0.1565 | No |
| 34 | Lsp1 | 4198 | 0.177 | 0.1557 | No |
| 35 | Eif4a2 | 4488 | 0.160 | 0.1432 | No |
| 36 | Actn3 | 4533 | 0.158 | 0.1454 | No |
| 37 | Crat | 4762 | 0.145 | 0.1361 | No |
| 38 | Atp2a1 | 4840 | 0.141 | 0.1358 | No |
| 39 | Fdps | 4909 | 0.137 | 0.1359 | No |
| 40 | Akt2 | 4987 | 0.133 | 0.1353 | No |
| 41 | Myh11 | 5303 | 0.116 | 0.1199 | No |
| 42 | Tsc2 | 5399 | 0.113 | 0.1176 | No |
| 43 | Ppfia4 | 5534 | 0.106 | 0.1128 | No |
| 44 | Cdkn1a | 5671 | 0.099 | 0.1076 | No |
| 45 | Mapre3 | 5675 | 0.099 | 0.1105 | No |
| 46 | Sh2b1 | 5781 | 0.094 | 0.1070 | No |
| 47 | Cdh13 | 5947 | 0.086 | 0.0997 | No |
| 48 | Pkia | 6020 | 0.082 | 0.0979 | No |
| 49 | Sphk1 | 6101 | 0.078 | 0.0954 | No |
| 50 | Slc6a8 | 6250 | 0.070 | 0.0887 | No |
| 51 | Mapk12 | 6361 | 0.065 | 0.0840 | No |
| 52 | Bhlhe40 | 6841 | 0.041 | 0.0564 | No |
| 53 | Chrnb1 | 7208 | 0.023 | 0.0350 | No |
| 54 | App | 7321 | 0.017 | 0.0287 | No |
| 55 | Lama2 | 7344 | 0.015 | 0.0279 | No |
| 56 | Erbb3 | 7442 | 0.011 | 0.0224 | No |
| 57 | Tead4 | 7486 | 0.010 | 0.0201 | No |
| 58 | Gaa | 7545 | 0.007 | 0.0168 | No |
| 59 | Fkbp1b | 7691 | 0.002 | 0.0081 | No |
| 60 | Mef2d | 7712 | 0.001 | 0.0069 | No |
| 61 | Col1a1 | 7734 | 0.000 | 0.0057 | No |
| 62 | Tnnc1 | 7759 | 0.000 | 0.0042 | No |
| 63 | Acsl1 | 7844 | -0.003 | -0.0008 | No |
| 64 | Psen2 | 7893 | -0.006 | -0.0035 | No |
| 65 | Ncam1 | 8098 | -0.016 | -0.0153 | No |
| 66 | Ryr1 | 8177 | -0.019 | -0.0194 | No |
| 67 | Igfbp7 | 8229 | -0.021 | -0.0218 | No |
| 68 | Ak1 | 8439 | -0.032 | -0.0334 | No |
| 69 | Wwtr1 | 8481 | -0.034 | -0.0349 | No |
| 70 | Adcy9 | 8612 | -0.040 | -0.0415 | No |
| 71 | Pdlim7 | 8759 | -0.046 | -0.0489 | No |
| 72 | Bin1 | 9417 | -0.079 | -0.0861 | No |
| 73 | Sorbs1 | 9917 | -0.104 | -0.1130 | No |
| 74 | Foxo4 | 9923 | -0.104 | -0.1101 | No |
| 75 | Svil | 10613 | -0.137 | -0.1474 | No |
| 76 | Pfkm | 10683 | -0.142 | -0.1472 | No |
| 77 | Tgfb1 | 11190 | -0.170 | -0.1726 | No |
| 78 | Fhl1 | 11419 | -0.181 | -0.1808 | No |
| 79 | Large1 | 11821 | -0.204 | -0.1987 | No |
| 80 | Myom1 | 11833 | -0.204 | -0.1931 | No |
| 81 | Kifc3 | 12005 | -0.214 | -0.1968 | No |
| 82 | Adam12 | 12081 | -0.218 | -0.1947 | No |
| 83 | Tpd52l1 | 12318 | -0.232 | -0.2018 | Yes |
| 84 | Tpm3 | 12350 | -0.234 | -0.1965 | Yes |
| 85 | Pcx | 12425 | -0.238 | -0.1937 | Yes |
| 86 | Myl4 | 12491 | -0.241 | -0.1902 | Yes |
| 87 | Ctf1 | 12576 | -0.245 | -0.1877 | Yes |
| 88 | Sirt2 | 12617 | -0.246 | -0.1826 | Yes |
| 89 | Actn2 | 12654 | -0.248 | -0.1771 | Yes |
| 90 | Csrp3 | 12674 | -0.249 | -0.1706 | Yes |
| 91 | Igf1 | 12790 | -0.258 | -0.1697 | Yes |
| 92 | Tpm2 | 13126 | -0.276 | -0.1814 | Yes |
| 93 | Gadd45b | 13143 | -0.278 | -0.1738 | Yes |
| 94 | Gpx3 | 13146 | -0.279 | -0.1654 | Yes |
| 95 | Ablim1 | 13226 | -0.285 | -0.1614 | Yes |
| 96 | Ache | 13620 | -0.315 | -0.1755 | Yes |
| 97 | Dmd | 13635 | -0.316 | -0.1667 | Yes |
| 98 | Prnp | 13696 | -0.321 | -0.1604 | Yes |
| 99 | Pygm | 13838 | -0.332 | -0.1588 | Yes |
| 100 | Eno3 | 14295 | -0.373 | -0.1748 | Yes |
| 101 | Sorbs3 | 14327 | -0.376 | -0.1652 | Yes |
| 102 | Kcnh2 | 14424 | -0.386 | -0.1591 | Yes |
| 103 | Cd36 | 14538 | -0.395 | -0.1538 | Yes |
| 104 | Mylk | 14691 | -0.414 | -0.1503 | Yes |
| 105 | Plxnb2 | 14718 | -0.417 | -0.1391 | Yes |
| 106 | Atp6ap1 | 14769 | -0.422 | -0.1292 | Yes |
| 107 | Nav2 | 14888 | -0.434 | -0.1230 | Yes |
| 108 | Fgf2 | 14941 | -0.439 | -0.1127 | Yes |
| 109 | Mybpc3 | 14947 | -0.440 | -0.0995 | Yes |
| 110 | Bag1 | 15175 | -0.464 | -0.0990 | Yes |
| 111 | Ptp4a3 | 15240 | -0.472 | -0.0883 | Yes |
| 112 | Syngr2 | 15321 | -0.482 | -0.0784 | Yes |
| 113 | Smtn | 15352 | -0.486 | -0.0653 | Yes |
| 114 | Sgcd | 15536 | -0.512 | -0.0606 | Yes |
| 115 | Schip1 | 15656 | -0.534 | -0.0514 | Yes |
| 116 | Mras | 15976 | -0.609 | -0.0520 | Yes |
| 117 | Nqo1 | 16137 | -0.654 | -0.0416 | Yes |
| 118 | Cryab | 16354 | -0.723 | -0.0324 | Yes |
| 119 | Cnn3 | 16459 | -0.790 | -0.0145 | Yes |
| 120 | Mef2c | 16609 | -0.942 | 0.0054 | Yes |