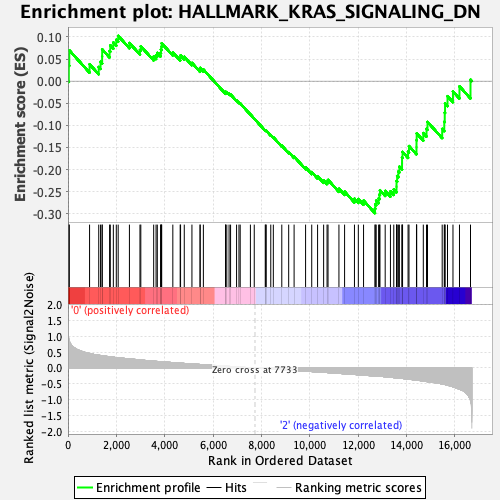
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.29870537 |
| Normalized Enrichment Score (NES) | -1.13453 |
| Nominal p-value | 0.24242425 |
| FDR q-value | 0.80201215 |
| FWER p-Value | 0.967 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nrip2 | 34 | 0.913 | 0.0357 | No |
| 2 | Ryr2 | 53 | 0.849 | 0.0697 | No |
| 3 | Macroh2a2 | 892 | 0.458 | 0.0382 | No |
| 4 | Thnsl2 | 1271 | 0.403 | 0.0321 | No |
| 5 | Epha5 | 1346 | 0.394 | 0.0439 | No |
| 6 | Ybx2 | 1412 | 0.386 | 0.0560 | No |
| 7 | Sgk1 | 1414 | 0.386 | 0.0719 | No |
| 8 | Stag3 | 1720 | 0.354 | 0.0681 | No |
| 9 | Thrb | 1749 | 0.351 | 0.0809 | No |
| 10 | Htr1b | 1875 | 0.339 | 0.0874 | No |
| 11 | Skil | 1997 | 0.329 | 0.0937 | No |
| 12 | Cyp39a1 | 2075 | 0.323 | 0.1025 | No |
| 13 | Nos1 | 2540 | 0.285 | 0.0863 | No |
| 14 | Btg2 | 2976 | 0.253 | 0.0706 | No |
| 15 | Entpd7 | 3011 | 0.250 | 0.0789 | No |
| 16 | Lfng | 3543 | 0.214 | 0.0557 | No |
| 17 | Vps50 | 3642 | 0.208 | 0.0584 | No |
| 18 | Grid2 | 3693 | 0.206 | 0.0639 | No |
| 19 | Zbtb16 | 3829 | 0.198 | 0.0640 | No |
| 20 | Zc2hc1c | 3843 | 0.197 | 0.0713 | No |
| 21 | Brdt | 3867 | 0.196 | 0.0780 | No |
| 22 | Celsr2 | 3874 | 0.195 | 0.0857 | No |
| 23 | Clstn3 | 4335 | 0.169 | 0.0650 | No |
| 24 | Kmt2d | 4639 | 0.152 | 0.0530 | No |
| 25 | Gp1ba | 4655 | 0.151 | 0.0583 | No |
| 26 | Copz2 | 4808 | 0.143 | 0.0551 | No |
| 27 | Tnni3 | 5125 | 0.125 | 0.0412 | No |
| 28 | Atp4a | 5461 | 0.110 | 0.0256 | No |
| 29 | Tg | 5465 | 0.110 | 0.0300 | No |
| 30 | Dtnb | 5598 | 0.103 | 0.0263 | No |
| 31 | Cacna1f | 6513 | 0.057 | -0.0264 | No |
| 32 | Zfp112 | 6514 | 0.057 | -0.0240 | No |
| 33 | Nr4a2 | 6566 | 0.054 | -0.0249 | No |
| 34 | Rsad2 | 6661 | 0.049 | -0.0285 | No |
| 35 | Cdkal1 | 6723 | 0.046 | -0.0303 | No |
| 36 | Mthfr | 6971 | 0.034 | -0.0438 | No |
| 37 | Msh5 | 7070 | 0.029 | -0.0484 | No |
| 38 | Sphk2 | 7121 | 0.027 | -0.0504 | No |
| 39 | Mefv | 7543 | 0.007 | -0.0754 | No |
| 40 | Kcnd1 | 7702 | 0.001 | -0.0849 | No |
| 41 | Arpp21 | 8157 | -0.018 | -0.1114 | No |
| 42 | Ryr1 | 8177 | -0.019 | -0.1118 | No |
| 43 | Capn9 | 8195 | -0.020 | -0.1120 | No |
| 44 | Snn | 8385 | -0.029 | -0.1222 | No |
| 45 | Itgb1bp2 | 8491 | -0.034 | -0.1271 | No |
| 46 | Efhd1 | 8841 | -0.050 | -0.1460 | No |
| 47 | Cd80 | 9126 | -0.064 | -0.1605 | No |
| 48 | Bard1 | 9350 | -0.076 | -0.1708 | No |
| 49 | Coq8a | 9825 | -0.099 | -0.1952 | No |
| 50 | Pdk2 | 10079 | -0.112 | -0.2058 | No |
| 51 | Prodh | 10321 | -0.124 | -0.2152 | No |
| 52 | Egf | 10571 | -0.135 | -0.2246 | No |
| 53 | Htr1d | 10713 | -0.143 | -0.2271 | No |
| 54 | Klk8 | 10753 | -0.146 | -0.2235 | No |
| 55 | Sptbn2 | 11204 | -0.171 | -0.2435 | No |
| 56 | Ypel1 | 11438 | -0.182 | -0.2500 | No |
| 57 | Tex15 | 11846 | -0.205 | -0.2660 | No |
| 58 | Slc29a3 | 12006 | -0.214 | -0.2667 | No |
| 59 | Gamt | 12222 | -0.227 | -0.2703 | No |
| 60 | Fggy | 12695 | -0.251 | -0.2883 | Yes |
| 61 | Mast3 | 12707 | -0.252 | -0.2786 | Yes |
| 62 | Sidt1 | 12745 | -0.255 | -0.2703 | Yes |
| 63 | Abcg4 | 12845 | -0.261 | -0.2654 | Yes |
| 64 | Cpb1 | 12879 | -0.263 | -0.2565 | Yes |
| 65 | Ngb | 12901 | -0.264 | -0.2469 | Yes |
| 66 | Ptprj | 13117 | -0.276 | -0.2484 | Yes |
| 67 | Asb7 | 13334 | -0.293 | -0.2493 | Yes |
| 68 | Magix | 13472 | -0.304 | -0.2450 | Yes |
| 69 | Myo15a | 13585 | -0.312 | -0.2389 | Yes |
| 70 | Tent5c | 13586 | -0.312 | -0.2260 | Yes |
| 71 | Selenop | 13613 | -0.314 | -0.2146 | Yes |
| 72 | Slc25a23 | 13662 | -0.318 | -0.2043 | Yes |
| 73 | Mfsd6 | 13704 | -0.322 | -0.1935 | Yes |
| 74 | Bmpr1b | 13814 | -0.330 | -0.1864 | Yes |
| 75 | Serpinb2 | 13815 | -0.330 | -0.1727 | Yes |
| 76 | Cpeb3 | 13835 | -0.331 | -0.1601 | Yes |
| 77 | Mx2 | 14063 | -0.350 | -0.1593 | Yes |
| 78 | Tcf7l1 | 14101 | -0.354 | -0.1469 | Yes |
| 79 | Idua | 14411 | -0.384 | -0.1497 | Yes |
| 80 | Tenm2 | 14412 | -0.384 | -0.1338 | Yes |
| 81 | Nr6a1 | 14420 | -0.385 | -0.1183 | Yes |
| 82 | Synpo | 14693 | -0.414 | -0.1175 | Yes |
| 83 | Gtf3c5 | 14826 | -0.428 | -0.1078 | Yes |
| 84 | Dcc | 14867 | -0.431 | -0.0924 | Yes |
| 85 | Tgm1 | 15474 | -0.503 | -0.1081 | Yes |
| 86 | Gpr19 | 15560 | -0.517 | -0.0918 | Yes |
| 87 | Tgfb2 | 15580 | -0.521 | -0.0714 | Yes |
| 88 | Camk1d | 15593 | -0.523 | -0.0505 | Yes |
| 89 | Ccdc106 | 15694 | -0.542 | -0.0341 | Yes |
| 90 | Prkn | 15920 | -0.593 | -0.0232 | Yes |
| 91 | Plag1 | 16189 | -0.666 | -0.0118 | Yes |
| 92 | Slc16a7 | 16645 | -1.026 | 0.0033 | Yes |

