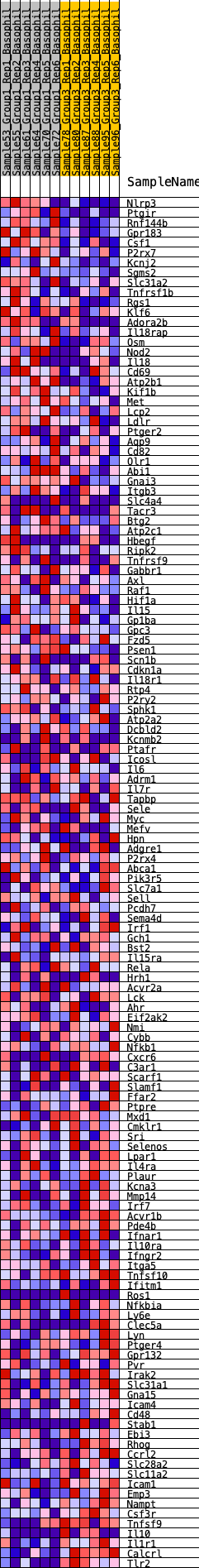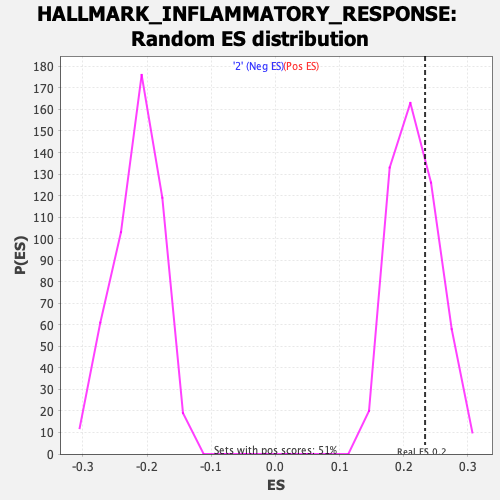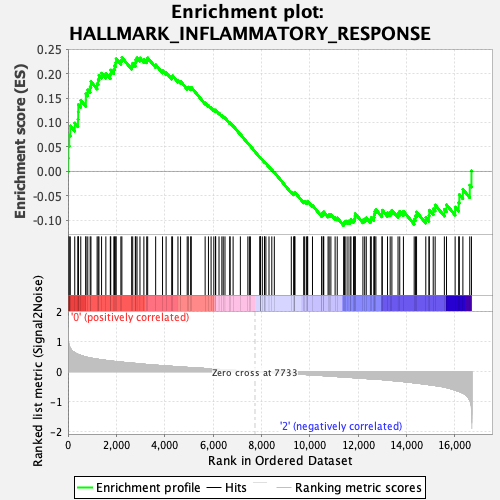
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.2331809 |
| Normalized Enrichment Score (NES) | 1.0769279 |
| Nominal p-value | 0.3392157 |
| FDR q-value | 0.5327527 |
| FWER p-Value | 0.982 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nlrp3 | 14 | 1.007 | 0.0266 | Yes |
| 2 | Ptgir | 18 | 0.979 | 0.0530 | Yes |
| 3 | Rnf144b | 52 | 0.850 | 0.0742 | Yes |
| 4 | Gpr183 | 94 | 0.779 | 0.0929 | Yes |
| 5 | Csf1 | 279 | 0.628 | 0.0989 | Yes |
| 6 | P2rx7 | 412 | 0.572 | 0.1065 | Yes |
| 7 | Kcnj2 | 422 | 0.569 | 0.1214 | Yes |
| 8 | Sgms2 | 427 | 0.567 | 0.1366 | Yes |
| 9 | Slc31a2 | 528 | 0.538 | 0.1452 | Yes |
| 10 | Tnfrsf1b | 738 | 0.481 | 0.1457 | Yes |
| 11 | Rgs1 | 739 | 0.481 | 0.1588 | Yes |
| 12 | Klf6 | 818 | 0.467 | 0.1668 | Yes |
| 13 | Adora2b | 917 | 0.454 | 0.1732 | Yes |
| 14 | Il18rap | 949 | 0.448 | 0.1836 | Yes |
| 15 | Osm | 1204 | 0.411 | 0.1794 | Yes |
| 16 | Nod2 | 1248 | 0.406 | 0.1879 | Yes |
| 17 | Il18 | 1280 | 0.402 | 0.1969 | Yes |
| 18 | Cd69 | 1388 | 0.388 | 0.2010 | Yes |
| 19 | Atp2b1 | 1565 | 0.370 | 0.2005 | Yes |
| 20 | Kif1b | 1750 | 0.351 | 0.1989 | Yes |
| 21 | Met | 1767 | 0.348 | 0.2074 | Yes |
| 22 | Lcp2 | 1897 | 0.337 | 0.2088 | Yes |
| 23 | Ldlr | 1925 | 0.335 | 0.2163 | Yes |
| 24 | Ptger2 | 1974 | 0.330 | 0.2224 | Yes |
| 25 | Aqp9 | 1989 | 0.329 | 0.2305 | Yes |
| 26 | Cd82 | 2187 | 0.313 | 0.2272 | Yes |
| 27 | Olr1 | 2228 | 0.310 | 0.2332 | Yes |
| 28 | Abi1 | 2633 | 0.279 | 0.2164 | No |
| 29 | Gnai3 | 2676 | 0.276 | 0.2214 | No |
| 30 | Itgb3 | 2784 | 0.269 | 0.2222 | No |
| 31 | Slc4a4 | 2801 | 0.268 | 0.2286 | No |
| 32 | Tacr3 | 2854 | 0.263 | 0.2326 | No |
| 33 | Btg2 | 2976 | 0.253 | 0.2322 | No |
| 34 | Atp2c1 | 3138 | 0.241 | 0.2290 | No |
| 35 | Hbegf | 3255 | 0.233 | 0.2284 | No |
| 36 | Ripk2 | 3295 | 0.231 | 0.2323 | No |
| 37 | Tnfrsf9 | 3625 | 0.209 | 0.2181 | No |
| 38 | Gabbr1 | 3909 | 0.193 | 0.2063 | No |
| 39 | Axl | 4057 | 0.184 | 0.2024 | No |
| 40 | Raf1 | 4289 | 0.171 | 0.1931 | No |
| 41 | Hif1a | 4324 | 0.169 | 0.1957 | No |
| 42 | Il15 | 4547 | 0.157 | 0.1866 | No |
| 43 | Gp1ba | 4655 | 0.151 | 0.1842 | No |
| 44 | Gpc3 | 4929 | 0.136 | 0.1714 | No |
| 45 | Fzd5 | 4973 | 0.134 | 0.1725 | No |
| 46 | Psen1 | 5072 | 0.128 | 0.1700 | No |
| 47 | Scn1b | 5097 | 0.127 | 0.1720 | No |
| 48 | Cdkn1a | 5671 | 0.099 | 0.1401 | No |
| 49 | Il18r1 | 5808 | 0.093 | 0.1344 | No |
| 50 | Rtp4 | 5917 | 0.088 | 0.1303 | No |
| 51 | P2ry2 | 6030 | 0.082 | 0.1258 | No |
| 52 | Sphk1 | 6101 | 0.078 | 0.1237 | No |
| 53 | Atp2a2 | 6106 | 0.078 | 0.1255 | No |
| 54 | Dcbld2 | 6245 | 0.070 | 0.1191 | No |
| 55 | Kcnmb2 | 6363 | 0.065 | 0.1138 | No |
| 56 | Ptafr | 6419 | 0.062 | 0.1122 | No |
| 57 | Icosl | 6492 | 0.058 | 0.1094 | No |
| 58 | Il6 | 6691 | 0.048 | 0.0987 | No |
| 59 | Adrm1 | 6703 | 0.047 | 0.0994 | No |
| 60 | Il7r | 6828 | 0.042 | 0.0930 | No |
| 61 | Tapbp | 7128 | 0.026 | 0.0757 | No |
| 62 | Sele | 7438 | 0.011 | 0.0573 | No |
| 63 | Myc | 7501 | 0.009 | 0.0538 | No |
| 64 | Mefv | 7543 | 0.007 | 0.0515 | No |
| 65 | Hpn | 7944 | -0.008 | 0.0276 | No |
| 66 | Adgre1 | 7946 | -0.008 | 0.0278 | No |
| 67 | P2rx4 | 7948 | -0.008 | 0.0279 | No |
| 68 | Abca1 | 7954 | -0.008 | 0.0279 | No |
| 69 | Pik3r5 | 8036 | -0.013 | 0.0233 | No |
| 70 | Slc7a1 | 8125 | -0.017 | 0.0185 | No |
| 71 | Sell | 8126 | -0.017 | 0.0189 | No |
| 72 | Pcdh7 | 8183 | -0.019 | 0.0161 | No |
| 73 | Sema4d | 8312 | -0.026 | 0.0091 | No |
| 74 | Irf1 | 8428 | -0.031 | 0.0030 | No |
| 75 | Gch1 | 8532 | -0.036 | -0.0023 | No |
| 76 | Bst2 | 9234 | -0.070 | -0.0427 | No |
| 77 | Il15ra | 9332 | -0.075 | -0.0465 | No |
| 78 | Rela | 9358 | -0.076 | -0.0459 | No |
| 79 | Hrh1 | 9377 | -0.077 | -0.0449 | No |
| 80 | Acvr2a | 9379 | -0.077 | -0.0429 | No |
| 81 | Lck | 9747 | -0.095 | -0.0624 | No |
| 82 | Ahr | 9786 | -0.098 | -0.0621 | No |
| 83 | Eif2ak2 | 9858 | -0.101 | -0.0636 | No |
| 84 | Nmi | 9899 | -0.103 | -0.0632 | No |
| 85 | Cybb | 9913 | -0.104 | -0.0612 | No |
| 86 | Nfkb1 | 10110 | -0.114 | -0.0699 | No |
| 87 | Cxcr6 | 10493 | -0.132 | -0.0894 | No |
| 88 | C3ar1 | 10496 | -0.132 | -0.0859 | No |
| 89 | Scarf1 | 10570 | -0.135 | -0.0867 | No |
| 90 | Slamf1 | 10573 | -0.135 | -0.0831 | No |
| 91 | Ffar2 | 10755 | -0.146 | -0.0900 | No |
| 92 | Ptpre | 10800 | -0.149 | -0.0886 | No |
| 93 | Mxd1 | 10871 | -0.152 | -0.0887 | No |
| 94 | Cmklr1 | 11046 | -0.163 | -0.0948 | No |
| 95 | Sri | 11134 | -0.167 | -0.0955 | No |
| 96 | Selenos | 11398 | -0.180 | -0.1065 | No |
| 97 | Lpar1 | 11436 | -0.182 | -0.1037 | No |
| 98 | Il4ra | 11491 | -0.185 | -0.1020 | No |
| 99 | Plaur | 11578 | -0.191 | -0.1020 | No |
| 100 | Kcna3 | 11666 | -0.195 | -0.1019 | No |
| 101 | Mmp14 | 11701 | -0.198 | -0.0986 | No |
| 102 | Irf7 | 11808 | -0.203 | -0.0995 | No |
| 103 | Acvr1b | 11855 | -0.205 | -0.0967 | No |
| 104 | Pde4b | 11868 | -0.206 | -0.0918 | No |
| 105 | Ifnar1 | 11877 | -0.206 | -0.0866 | No |
| 106 | Il10ra | 12196 | -0.226 | -0.0997 | No |
| 107 | Ifngr2 | 12269 | -0.230 | -0.0978 | No |
| 108 | Itga5 | 12334 | -0.233 | -0.0953 | No |
| 109 | Tnfsf10 | 12505 | -0.241 | -0.0990 | No |
| 110 | Ifitm1 | 12542 | -0.244 | -0.0946 | No |
| 111 | Ros1 | 12650 | -0.248 | -0.0942 | No |
| 112 | Nfkbia | 12665 | -0.249 | -0.0883 | No |
| 113 | Ly6e | 12686 | -0.251 | -0.0827 | No |
| 114 | Clec5a | 12733 | -0.254 | -0.0786 | No |
| 115 | Lyn | 12984 | -0.267 | -0.0864 | No |
| 116 | Ptger4 | 12995 | -0.268 | -0.0797 | No |
| 117 | Gpr132 | 13213 | -0.284 | -0.0851 | No |
| 118 | Pvr | 13323 | -0.292 | -0.0837 | No |
| 119 | Irak2 | 13394 | -0.297 | -0.0798 | No |
| 120 | Slc31a1 | 13648 | -0.316 | -0.0865 | No |
| 121 | Gna15 | 13719 | -0.324 | -0.0819 | No |
| 122 | Icam4 | 13871 | -0.334 | -0.0819 | No |
| 123 | Cd48 | 14316 | -0.375 | -0.0985 | No |
| 124 | Stab1 | 14368 | -0.381 | -0.0912 | No |
| 125 | Ebi3 | 14414 | -0.384 | -0.0835 | No |
| 126 | Rhog | 14796 | -0.425 | -0.0949 | No |
| 127 | Ccrl2 | 14927 | -0.437 | -0.0908 | No |
| 128 | Slc28a2 | 14940 | -0.438 | -0.0796 | No |
| 129 | Slc11a2 | 15103 | -0.457 | -0.0770 | No |
| 130 | Icam1 | 15183 | -0.466 | -0.0691 | No |
| 131 | Emp3 | 15564 | -0.518 | -0.0779 | No |
| 132 | Nampt | 15653 | -0.534 | -0.0687 | No |
| 133 | Csf3r | 16009 | -0.618 | -0.0733 | No |
| 134 | Tnfsf9 | 16157 | -0.658 | -0.0643 | No |
| 135 | Il10 | 16183 | -0.664 | -0.0477 | No |
| 136 | Il1r1 | 16330 | -0.713 | -0.0371 | No |
| 137 | Calcrl | 16614 | -0.948 | -0.0284 | No |
| 138 | Tlr2 | 16684 | -1.228 | 0.0009 | No |