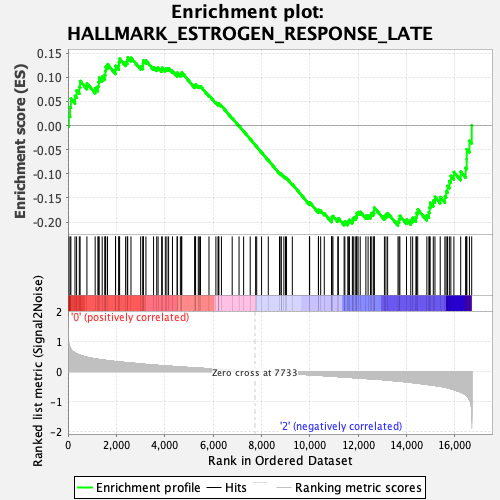
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | -0.2071662 |
| Normalized Enrichment Score (NES) | -0.953197 |
| Nominal p-value | 0.5913371 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdh1 | 35 | 0.902 | 0.0211 | No |
| 2 | Ptges | 87 | 0.793 | 0.0384 | No |
| 3 | Unc13b | 119 | 0.742 | 0.0557 | No |
| 4 | Jak2 | 289 | 0.622 | 0.0615 | No |
| 5 | Fos | 355 | 0.590 | 0.0728 | No |
| 6 | Sfn | 468 | 0.553 | 0.0802 | No |
| 7 | Btg3 | 500 | 0.546 | 0.0924 | No |
| 8 | Jak1 | 782 | 0.474 | 0.0877 | No |
| 9 | Tiam1 | 1123 | 0.420 | 0.0779 | No |
| 10 | Ptger3 | 1236 | 0.408 | 0.0817 | No |
| 11 | Aff1 | 1267 | 0.403 | 0.0902 | No |
| 12 | Klf4 | 1288 | 0.400 | 0.0993 | No |
| 13 | Sgk1 | 1414 | 0.386 | 0.1017 | No |
| 14 | Plxnb1 | 1522 | 0.375 | 0.1049 | No |
| 15 | Tjp3 | 1545 | 0.373 | 0.1132 | No |
| 16 | Papss2 | 1560 | 0.371 | 0.1219 | No |
| 17 | Rps6ka2 | 1634 | 0.363 | 0.1269 | No |
| 18 | Slc16a1 | 1964 | 0.331 | 0.1155 | No |
| 19 | Mest | 1968 | 0.331 | 0.1238 | No |
| 20 | Siah2 | 2099 | 0.321 | 0.1242 | No |
| 21 | Isg20 | 2108 | 0.320 | 0.1320 | No |
| 22 | Homer2 | 2135 | 0.317 | 0.1386 | No |
| 23 | Id2 | 2383 | 0.297 | 0.1313 | No |
| 24 | Abhd2 | 2449 | 0.293 | 0.1350 | No |
| 25 | Stil | 2469 | 0.290 | 0.1413 | No |
| 26 | Impa2 | 2604 | 0.282 | 0.1404 | No |
| 27 | Flnb | 3005 | 0.250 | 0.1227 | No |
| 28 | Cdc6 | 3099 | 0.243 | 0.1234 | No |
| 29 | Cd9 | 3102 | 0.243 | 0.1295 | No |
| 30 | Cav1 | 3116 | 0.242 | 0.1350 | No |
| 31 | Gfus | 3223 | 0.235 | 0.1346 | No |
| 32 | Xbp1 | 3538 | 0.215 | 0.1212 | No |
| 33 | Dhcr7 | 3669 | 0.206 | 0.1186 | No |
| 34 | Il17rb | 3725 | 0.203 | 0.1206 | No |
| 35 | Celsr2 | 3874 | 0.195 | 0.1166 | No |
| 36 | Top2a | 3899 | 0.194 | 0.1202 | No |
| 37 | Slc24a3 | 4025 | 0.186 | 0.1174 | No |
| 38 | Amfr | 4090 | 0.182 | 0.1182 | No |
| 39 | Cacna2d2 | 4160 | 0.178 | 0.1187 | No |
| 40 | Pgr | 4320 | 0.170 | 0.1134 | No |
| 41 | Dnajc1 | 4515 | 0.159 | 0.1058 | No |
| 42 | Ncor2 | 4518 | 0.159 | 0.1098 | No |
| 43 | Slc1a4 | 4647 | 0.151 | 0.1059 | No |
| 44 | Scarb1 | 4698 | 0.148 | 0.1067 | No |
| 45 | St14 | 4702 | 0.148 | 0.1104 | No |
| 46 | Bcl2 | 5238 | 0.119 | 0.0811 | No |
| 47 | Abca3 | 5258 | 0.118 | 0.0830 | No |
| 48 | Add3 | 5276 | 0.117 | 0.0850 | No |
| 49 | Slc7a5 | 5388 | 0.114 | 0.0812 | No |
| 50 | Gale | 5431 | 0.112 | 0.0816 | No |
| 51 | Mapt | 5479 | 0.110 | 0.0815 | No |
| 52 | Rbbp8 | 5830 | 0.091 | 0.0628 | No |
| 53 | Aldh3a2 | 6116 | 0.077 | 0.0475 | No |
| 54 | Rab31 | 6207 | 0.073 | 0.0440 | No |
| 55 | Gla | 6218 | 0.072 | 0.0452 | No |
| 56 | Olfm1 | 6239 | 0.071 | 0.0459 | No |
| 57 | Pdcd4 | 6345 | 0.066 | 0.0412 | No |
| 58 | Perp | 6790 | 0.043 | 0.0155 | No |
| 59 | Kif20a | 7073 | 0.029 | -0.0008 | No |
| 60 | Mettl3 | 7263 | 0.020 | -0.0116 | No |
| 61 | Hspa4l | 7535 | 0.008 | -0.0278 | No |
| 62 | Tnnc1 | 7759 | 0.000 | -0.0413 | No |
| 63 | Slc22a5 | 7803 | -0.002 | -0.0438 | No |
| 64 | Ugdh | 8005 | -0.011 | -0.0557 | No |
| 65 | Nab2 | 8282 | -0.025 | -0.0717 | No |
| 66 | Snx10 | 8746 | -0.046 | -0.0985 | No |
| 67 | Farp1 | 8811 | -0.049 | -0.1011 | No |
| 68 | Fkbp4 | 8815 | -0.049 | -0.1000 | No |
| 69 | Fabp5 | 8927 | -0.054 | -0.1053 | No |
| 70 | Idh2 | 8994 | -0.058 | -0.1078 | No |
| 71 | Tob1 | 9037 | -0.059 | -0.1088 | No |
| 72 | Prkar2b | 9276 | -0.072 | -0.1213 | No |
| 73 | Blvrb | 9984 | -0.107 | -0.1612 | No |
| 74 | Tst | 9993 | -0.108 | -0.1589 | No |
| 75 | Etfb | 10356 | -0.126 | -0.1776 | No |
| 76 | Cish | 10357 | -0.126 | -0.1743 | No |
| 77 | Tspan13 | 10449 | -0.130 | -0.1765 | No |
| 78 | Rnaseh2a | 10598 | -0.137 | -0.1819 | No |
| 79 | Myb | 10896 | -0.154 | -0.1959 | No |
| 80 | Wfs1 | 10904 | -0.154 | -0.1923 | No |
| 81 | Llgl2 | 10916 | -0.155 | -0.1890 | No |
| 82 | Fkbp5 | 10956 | -0.157 | -0.1873 | No |
| 83 | Pdzk1 | 11158 | -0.168 | -0.1951 | No |
| 84 | Mocs2 | 11180 | -0.169 | -0.1920 | No |
| 85 | Clic3 | 11418 | -0.181 | -0.2016 | Yes |
| 86 | Tmprss3 | 11449 | -0.183 | -0.1988 | Yes |
| 87 | Hmgcs2 | 11573 | -0.190 | -0.2013 | Yes |
| 88 | Ptpn6 | 11593 | -0.191 | -0.1975 | Yes |
| 89 | Dynlt3 | 11639 | -0.194 | -0.1952 | Yes |
| 90 | S100a9 | 11759 | -0.201 | -0.1973 | Yes |
| 91 | Scnn1a | 11781 | -0.201 | -0.1933 | Yes |
| 92 | Large1 | 11821 | -0.204 | -0.1905 | Yes |
| 93 | Dnajc12 | 11900 | -0.207 | -0.1898 | Yes |
| 94 | Slc29a1 | 11936 | -0.210 | -0.1865 | Yes |
| 95 | Sult2b1 | 11941 | -0.211 | -0.1813 | Yes |
| 96 | Elovl5 | 11996 | -0.214 | -0.1791 | Yes |
| 97 | Plk4 | 12084 | -0.219 | -0.1787 | Yes |
| 98 | Tpd52l1 | 12318 | -0.232 | -0.1868 | Yes |
| 99 | Igfbp4 | 12410 | -0.238 | -0.1862 | Yes |
| 100 | Arl3 | 12508 | -0.242 | -0.1858 | Yes |
| 101 | Anxa9 | 12540 | -0.244 | -0.1814 | Yes |
| 102 | Myof | 12622 | -0.247 | -0.1800 | Yes |
| 103 | St6galnac2 | 12659 | -0.248 | -0.1758 | Yes |
| 104 | Rabep1 | 12666 | -0.249 | -0.1697 | Yes |
| 105 | Nrip1 | 13085 | -0.274 | -0.1879 | Yes |
| 106 | Itpk1 | 13148 | -0.279 | -0.1845 | Yes |
| 107 | Zfp36 | 13218 | -0.284 | -0.1813 | Yes |
| 108 | Car12 | 13647 | -0.316 | -0.1990 | Yes |
| 109 | Atp2b4 | 13690 | -0.321 | -0.1933 | Yes |
| 110 | Gins2 | 13724 | -0.324 | -0.1870 | Yes |
| 111 | Opn3 | 13999 | -0.345 | -0.1946 | Yes |
| 112 | Dusp2 | 14175 | -0.361 | -0.1959 | Yes |
| 113 | Fdft1 | 14253 | -0.369 | -0.1910 | Yes |
| 114 | Cd44 | 14387 | -0.382 | -0.1892 | Yes |
| 115 | Batf | 14415 | -0.384 | -0.1810 | Yes |
| 116 | Lsr | 14462 | -0.388 | -0.1738 | Yes |
| 117 | Dcxr | 14840 | -0.430 | -0.1855 | Yes |
| 118 | Slc2a8 | 14923 | -0.437 | -0.1792 | Yes |
| 119 | Xrcc3 | 14950 | -0.440 | -0.1694 | Yes |
| 120 | Cox6c | 14977 | -0.443 | -0.1596 | Yes |
| 121 | Ppif | 15106 | -0.457 | -0.1556 | Yes |
| 122 | Bag1 | 15175 | -0.464 | -0.1477 | Yes |
| 123 | Chpt1 | 15392 | -0.492 | -0.1481 | Yes |
| 124 | Il6st | 15586 | -0.522 | -0.1463 | Yes |
| 125 | Slc26a2 | 15638 | -0.530 | -0.1357 | Yes |
| 126 | Ccnd1 | 15695 | -0.543 | -0.1251 | Yes |
| 127 | Cdc20 | 15770 | -0.559 | -0.1152 | Yes |
| 128 | Plaat3 | 15832 | -0.572 | -0.1042 | Yes |
| 129 | Nxt1 | 15958 | -0.605 | -0.0962 | Yes |
| 130 | Kcnk5 | 16239 | -0.686 | -0.0954 | Yes |
| 131 | Rapgefl1 | 16443 | -0.777 | -0.0877 | Yes |
| 132 | Sord | 16487 | -0.806 | -0.0695 | Yes |
| 133 | Car2 | 16490 | -0.810 | -0.0488 | Yes |
| 134 | Pkp3 | 16601 | -0.935 | -0.0314 | Yes |
| 135 | Ass1 | 16694 | -1.447 | 0.0003 | Yes |

