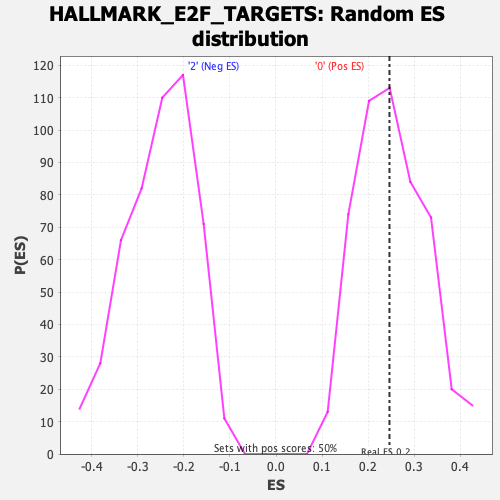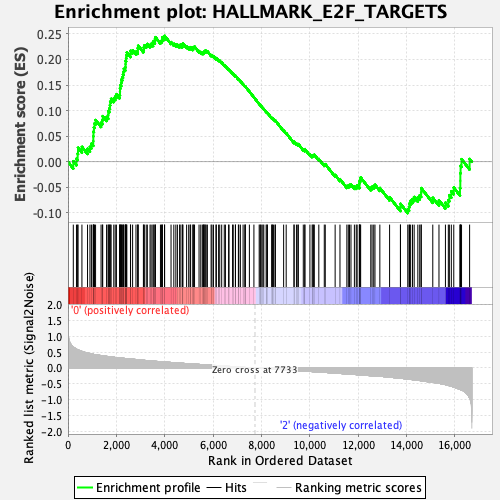
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group1_versus_Group3.Basophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_E2F_TARGETS |
| Enrichment Score (ES) | 0.2460374 |
| Normalized Enrichment Score (NES) | 0.9821877 |
| Nominal p-value | 0.46506986 |
| FDR q-value | 0.6058457 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rrm2 | 219 | 0.659 | 0.0011 | Yes |
| 2 | Zw10 | 350 | 0.592 | 0.0062 | Yes |
| 3 | Hnrnpd | 399 | 0.575 | 0.0159 | Yes |
| 4 | Lbr | 414 | 0.571 | 0.0275 | Yes |
| 5 | Lmnb1 | 576 | 0.523 | 0.0292 | Yes |
| 6 | Tcf19 | 808 | 0.468 | 0.0255 | Yes |
| 7 | Asf1b | 908 | 0.455 | 0.0294 | Yes |
| 8 | Donson | 968 | 0.445 | 0.0356 | Yes |
| 9 | Ddx39a | 1044 | 0.433 | 0.0405 | Yes |
| 10 | Nbn | 1047 | 0.432 | 0.0498 | Yes |
| 11 | Mcm4 | 1052 | 0.431 | 0.0590 | Yes |
| 12 | Cdkn2a | 1072 | 0.427 | 0.0672 | Yes |
| 13 | Brms1l | 1097 | 0.424 | 0.0750 | Yes |
| 14 | Anp32e | 1140 | 0.418 | 0.0816 | Yes |
| 15 | Rad21 | 1367 | 0.391 | 0.0765 | Yes |
| 16 | Dnmt1 | 1423 | 0.385 | 0.0816 | Yes |
| 17 | Brca1 | 1436 | 0.384 | 0.0892 | Yes |
| 18 | Psmc3ip | 1595 | 0.367 | 0.0877 | Yes |
| 19 | Dlgap5 | 1662 | 0.360 | 0.0916 | Yes |
| 20 | Pan2 | 1665 | 0.360 | 0.0993 | Yes |
| 21 | Spag5 | 1710 | 0.355 | 0.1044 | Yes |
| 22 | Melk | 1735 | 0.352 | 0.1107 | Yes |
| 23 | Syncrip | 1746 | 0.351 | 0.1177 | Yes |
| 24 | Atad2 | 1783 | 0.347 | 0.1231 | Yes |
| 25 | Msh2 | 1893 | 0.337 | 0.1239 | Yes |
| 26 | Timeless | 1952 | 0.332 | 0.1276 | Yes |
| 27 | Chek1 | 2002 | 0.328 | 0.1319 | Yes |
| 28 | Smc3 | 2133 | 0.318 | 0.1309 | Yes |
| 29 | Pole | 2149 | 0.316 | 0.1369 | Yes |
| 30 | Suv39h1 | 2151 | 0.316 | 0.1438 | Yes |
| 31 | Racgap1 | 2168 | 0.315 | 0.1497 | Yes |
| 32 | Mcm7 | 2200 | 0.312 | 0.1546 | Yes |
| 33 | Asf1a | 2210 | 0.311 | 0.1609 | Yes |
| 34 | Mre11a | 2247 | 0.308 | 0.1655 | Yes |
| 35 | Cdc25a | 2274 | 0.306 | 0.1706 | Yes |
| 36 | Mcm3 | 2298 | 0.305 | 0.1759 | Yes |
| 37 | Mthfd2 | 2312 | 0.303 | 0.1817 | Yes |
| 38 | Rbbp7 | 2372 | 0.298 | 0.1846 | Yes |
| 39 | Mcm2 | 2380 | 0.297 | 0.1907 | Yes |
| 40 | Pcna | 2382 | 0.297 | 0.1972 | Yes |
| 41 | Mcm6 | 2404 | 0.296 | 0.2024 | Yes |
| 42 | Kif22 | 2415 | 0.295 | 0.2082 | Yes |
| 43 | Smc1a | 2433 | 0.294 | 0.2136 | Yes |
| 44 | Cdk1 | 2584 | 0.283 | 0.2107 | Yes |
| 45 | Xpo1 | 2587 | 0.283 | 0.2168 | Yes |
| 46 | Ppm1d | 2672 | 0.276 | 0.2177 | Yes |
| 47 | Pds5b | 2811 | 0.267 | 0.2152 | Yes |
| 48 | Cse1l | 2886 | 0.260 | 0.2164 | Yes |
| 49 | Rad51ap1 | 2887 | 0.260 | 0.2221 | Yes |
| 50 | Ccp110 | 2902 | 0.258 | 0.2269 | Yes |
| 51 | Mlh1 | 3118 | 0.242 | 0.2192 | Yes |
| 52 | Cit | 3133 | 0.241 | 0.2236 | Yes |
| 53 | Pold3 | 3156 | 0.240 | 0.2275 | Yes |
| 54 | Cdc25b | 3234 | 0.234 | 0.2280 | Yes |
| 55 | Eif2s1 | 3281 | 0.232 | 0.2303 | Yes |
| 56 | Mms22l | 3393 | 0.224 | 0.2284 | Yes |
| 57 | Mad2l1 | 3438 | 0.221 | 0.2306 | Yes |
| 58 | Cdkn2c | 3518 | 0.216 | 0.2305 | Yes |
| 59 | Nup153 | 3521 | 0.216 | 0.2351 | Yes |
| 60 | Slbp | 3594 | 0.211 | 0.2354 | Yes |
| 61 | Ubr7 | 3598 | 0.211 | 0.2398 | Yes |
| 62 | Nup107 | 3615 | 0.210 | 0.2434 | Yes |
| 63 | Usp1 | 3823 | 0.198 | 0.2352 | Yes |
| 64 | Dctpp1 | 3869 | 0.195 | 0.2368 | Yes |
| 65 | Top2a | 3899 | 0.194 | 0.2393 | Yes |
| 66 | Ccne1 | 3900 | 0.194 | 0.2435 | Yes |
| 67 | Wee1 | 3991 | 0.188 | 0.2422 | Yes |
| 68 | Cdca8 | 3996 | 0.188 | 0.2460 | Yes |
| 69 | Pnn | 4261 | 0.173 | 0.2338 | No |
| 70 | Hells | 4376 | 0.166 | 0.2306 | No |
| 71 | Hmmr | 4460 | 0.161 | 0.2291 | No |
| 72 | Pola2 | 4524 | 0.158 | 0.2287 | No |
| 73 | Eed | 4616 | 0.153 | 0.2265 | No |
| 74 | Rad1 | 4637 | 0.152 | 0.2286 | No |
| 75 | Stag1 | 4724 | 0.147 | 0.2266 | No |
| 76 | Diaph3 | 4727 | 0.146 | 0.2297 | No |
| 77 | Nolc1 | 4754 | 0.145 | 0.2313 | No |
| 78 | Cenpe | 4905 | 0.138 | 0.2252 | No |
| 79 | Prkdc | 4991 | 0.133 | 0.2230 | No |
| 80 | Tmpo | 5049 | 0.130 | 0.2224 | No |
| 81 | Rfc1 | 5071 | 0.128 | 0.2239 | No |
| 82 | Kif2c | 5166 | 0.123 | 0.2209 | No |
| 83 | Kif18b | 5171 | 0.123 | 0.2234 | No |
| 84 | Aurkb | 5201 | 0.121 | 0.2242 | No |
| 85 | Smc4 | 5233 | 0.119 | 0.2250 | No |
| 86 | Espl1 | 5419 | 0.112 | 0.2162 | No |
| 87 | Depdc1a | 5491 | 0.109 | 0.2143 | No |
| 88 | Smc6 | 5555 | 0.106 | 0.2128 | No |
| 89 | Nasp | 5595 | 0.104 | 0.2127 | No |
| 90 | Aurka | 5610 | 0.103 | 0.2141 | No |
| 91 | Lig1 | 5626 | 0.102 | 0.2154 | No |
| 92 | Ncapd2 | 5664 | 0.099 | 0.2154 | No |
| 93 | Cdkn1a | 5671 | 0.099 | 0.2172 | No |
| 94 | Mki67 | 5715 | 0.097 | 0.2167 | No |
| 95 | Ilf3 | 5764 | 0.095 | 0.2158 | No |
| 96 | Dut | 5918 | 0.088 | 0.2085 | No |
| 97 | Wdr90 | 5954 | 0.086 | 0.2082 | No |
| 98 | Gspt1 | 6015 | 0.082 | 0.2064 | No |
| 99 | Rad51c | 6118 | 0.077 | 0.2019 | No |
| 100 | Kif4 | 6135 | 0.077 | 0.2026 | No |
| 101 | Srsf2 | 6228 | 0.072 | 0.1986 | No |
| 102 | Rpa2 | 6275 | 0.068 | 0.1973 | No |
| 103 | Tra2b | 6356 | 0.065 | 0.1939 | No |
| 104 | Mybl2 | 6462 | 0.059 | 0.1888 | No |
| 105 | Ipo7 | 6510 | 0.057 | 0.1872 | No |
| 106 | Tbrg4 | 6642 | 0.050 | 0.1804 | No |
| 107 | Plk1 | 6658 | 0.049 | 0.1805 | No |
| 108 | Lyar | 6805 | 0.043 | 0.1726 | No |
| 109 | Prim2 | 6832 | 0.041 | 0.1720 | No |
| 110 | Prdx4 | 6923 | 0.037 | 0.1673 | No |
| 111 | Cnot9 | 6924 | 0.037 | 0.1681 | No |
| 112 | Gins3 | 7046 | 0.030 | 0.1615 | No |
| 113 | Nme1 | 7064 | 0.029 | 0.1611 | No |
| 114 | Dek | 7129 | 0.026 | 0.1578 | No |
| 115 | Dscc1 | 7240 | 0.021 | 0.1516 | No |
| 116 | Nap1l1 | 7318 | 0.017 | 0.1473 | No |
| 117 | E2f8 | 7326 | 0.017 | 0.1472 | No |
| 118 | Ssrp1 | 7331 | 0.016 | 0.1473 | No |
| 119 | Myc | 7501 | 0.009 | 0.1373 | No |
| 120 | Ak2 | 7688 | 0.002 | 0.1260 | No |
| 121 | Cks2 | 7906 | -0.006 | 0.1130 | No |
| 122 | Cdkn1b | 7942 | -0.008 | 0.1111 | No |
| 123 | Hmgb2 | 7967 | -0.009 | 0.1098 | No |
| 124 | Tubg1 | 8015 | -0.012 | 0.1072 | No |
| 125 | Bub1b | 8060 | -0.014 | 0.1049 | No |
| 126 | Tubb5 | 8097 | -0.016 | 0.1030 | No |
| 127 | Cbx5 | 8194 | -0.020 | 0.0977 | No |
| 128 | Luc7l3 | 8226 | -0.021 | 0.0963 | No |
| 129 | Dclre1b | 8248 | -0.023 | 0.0955 | No |
| 130 | Mcm5 | 8424 | -0.031 | 0.0856 | No |
| 131 | Tacc3 | 8443 | -0.032 | 0.0852 | No |
| 132 | Rpa1 | 8451 | -0.033 | 0.0855 | No |
| 133 | Cdca3 | 8501 | -0.035 | 0.0832 | No |
| 134 | Ccnb2 | 8561 | -0.038 | 0.0805 | No |
| 135 | Srsf1 | 8577 | -0.038 | 0.0804 | No |
| 136 | Ctcf | 8917 | -0.054 | 0.0611 | No |
| 137 | Rad50 | 9023 | -0.059 | 0.0560 | No |
| 138 | Psip1 | 9346 | -0.076 | 0.0381 | No |
| 139 | Bard1 | 9350 | -0.076 | 0.0396 | No |
| 140 | Shmt1 | 9447 | -0.080 | 0.0355 | No |
| 141 | Pa2g4 | 9505 | -0.083 | 0.0339 | No |
| 142 | Xrcc6 | 9525 | -0.084 | 0.0346 | No |
| 143 | Rpa3 | 9730 | -0.094 | 0.0243 | No |
| 144 | Tipin | 9792 | -0.098 | 0.0227 | No |
| 145 | Brca2 | 9796 | -0.098 | 0.0247 | No |
| 146 | Nudt21 | 10004 | -0.108 | 0.0145 | No |
| 147 | Ezh2 | 10096 | -0.113 | 0.0115 | No |
| 148 | Pold2 | 10126 | -0.115 | 0.0123 | No |
| 149 | Trp53 | 10157 | -0.116 | 0.0130 | No |
| 150 | Orc2 | 10185 | -0.118 | 0.0139 | No |
| 151 | H2az1 | 10372 | -0.127 | 0.0054 | No |
| 152 | Rnaseh2a | 10598 | -0.137 | -0.0052 | No |
| 153 | Spc25 | 10639 | -0.139 | -0.0046 | No |
| 154 | Prps1 | 11047 | -0.163 | -0.0257 | No |
| 155 | Jpt1 | 11247 | -0.173 | -0.0340 | No |
| 156 | Pttg1 | 11532 | -0.188 | -0.0471 | No |
| 157 | Nup205 | 11592 | -0.191 | -0.0465 | No |
| 158 | Trip13 | 11640 | -0.194 | -0.0451 | No |
| 159 | Pold1 | 11702 | -0.198 | -0.0444 | No |
| 160 | Pms2 | 11843 | -0.205 | -0.0484 | No |
| 161 | Rfc2 | 11919 | -0.209 | -0.0484 | No |
| 162 | Stmn1 | 11957 | -0.212 | -0.0460 | No |
| 163 | Exosc8 | 12049 | -0.216 | -0.0468 | No |
| 164 | Ppp1r8 | 12050 | -0.216 | -0.0421 | No |
| 165 | Cks1b | 12058 | -0.217 | -0.0377 | No |
| 166 | Plk4 | 12084 | -0.219 | -0.0345 | No |
| 167 | Tfrc | 12102 | -0.220 | -0.0307 | No |
| 168 | Paics | 12521 | -0.243 | -0.0507 | No |
| 169 | Ranbp1 | 12574 | -0.245 | -0.0485 | No |
| 170 | Lsm8 | 12643 | -0.248 | -0.0472 | No |
| 171 | Hmga1b | 12694 | -0.251 | -0.0447 | No |
| 172 | H2ax | 12896 | -0.264 | -0.0512 | No |
| 173 | Nop56 | 13299 | -0.291 | -0.0692 | No |
| 174 | Cdkn3 | 13746 | -0.326 | -0.0891 | No |
| 175 | Chek2 | 13750 | -0.326 | -0.0821 | No |
| 176 | Mxd3 | 14051 | -0.349 | -0.0926 | No |
| 177 | Tk1 | 14104 | -0.354 | -0.0881 | No |
| 178 | Ran | 14122 | -0.355 | -0.0813 | No |
| 179 | Phf5a | 14168 | -0.360 | -0.0762 | No |
| 180 | Dck | 14245 | -0.368 | -0.0727 | No |
| 181 | Hmgb3 | 14315 | -0.375 | -0.0687 | No |
| 182 | Pop7 | 14467 | -0.388 | -0.0693 | No |
| 183 | Spc24 | 14545 | -0.396 | -0.0654 | No |
| 184 | Cdk4 | 14604 | -0.404 | -0.0600 | No |
| 185 | Rfc3 | 14607 | -0.404 | -0.0513 | No |
| 186 | Hus1 | 15085 | -0.455 | -0.0703 | No |
| 187 | Snrpb | 15341 | -0.484 | -0.0751 | No |
| 188 | Gins4 | 15608 | -0.526 | -0.0797 | No |
| 189 | Ing3 | 15728 | -0.548 | -0.0750 | No |
| 190 | Cdc20 | 15770 | -0.559 | -0.0652 | No |
| 191 | Gins1 | 15852 | -0.576 | -0.0575 | No |
| 192 | Ube2s | 15953 | -0.603 | -0.0504 | No |
| 193 | Orc6 | 16210 | -0.673 | -0.0512 | No |
| 194 | Ube2t | 16224 | -0.678 | -0.0371 | No |
| 195 | Ung | 16225 | -0.679 | -0.0223 | No |
| 196 | Birc5 | 16235 | -0.684 | -0.0079 | No |
| 197 | Pole4 | 16274 | -0.698 | 0.0051 | No |
| 198 | Cenpm | 16608 | -0.941 | 0.0055 | No |